3DB8
 
 | | Crystal structure of an activated (Thr->Asp) Polo-like kinase 1 (Plk1) catalytic domain in complex with Compound 041 | | Descriptor: | 3-[3-chloro-5-(5-{[(1S)-1-phenylethyl]amino}isoxazolo[5,4-c]pyridin-3-yl)phenyl]propan-1-ol, Polo-like kinase 1 | | Authors: | Elling, R.A, Hanan, E.J, Lew, W, Romanowski, M.J. | | Deposit date: | 2008-05-30 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Design and synthesis of 2-amino-isoxazolopyridines as Polo-like kinase inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
7S9E
 
 | | Cryo-EM Structure of dolphin Prestin: Inhibited II (Sulfate +Salicylate) state | | Descriptor: | 2-HYDROXYBENZOIC ACID, Prestin | | Authors: | Bavi, N, Clark, M.D, Contreras, G.F, Shen, R, Reddy, B.G, Milewski, W, Perozo, E. | | Deposit date: | 2021-09-20 | | Release date: | 2021-11-03 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The conformational cycle of prestin underlies outer-hair cell electromotility.
Nature, 600, 2021
|
|
7S8X
 
 | | Cryo-EM Structure of dolphin Prestin: Sensor Up (compact) state | | Descriptor: | Prestin | | Authors: | Bavi, N, Clark, M.D, Contreras, G.F, Shen, R, Reddy, B.G, Milewski, W, Perozo, E. | | Deposit date: | 2021-09-20 | | Release date: | 2021-11-03 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The conformational cycle of prestin underlies outer-hair cell electromotility.
Nature, 600, 2021
|
|
7S9C
 
 | | Cryo-EM Structure of dolphin Prestin: Sensor Down II (Expanded II) state | | Descriptor: | Prestin | | Authors: | Bavi, N, Clark, M.D, Contreras, G.F, Shen, R, Reddy, B.G, Milewski, W, Perozo, E. | | Deposit date: | 2021-09-20 | | Release date: | 2021-11-03 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | The conformational cycle of prestin underlies outer-hair cell electromotility.
Nature, 600, 2021
|
|
7S9D
 
 | | Cryo-EM Structure of dolphin Prestin: Intermediate state | | Descriptor: | Prestin | | Authors: | Bavi, N, Clark, M.D, Contreras, G.F, Shen, R, Reddy, B.G, Milewski, W, Perozo, E. | | Deposit date: | 2021-09-20 | | Release date: | 2021-11-03 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | The conformational cycle of prestin underlies outer-hair cell electromotility.
Nature, 600, 2021
|
|
7S9A
 
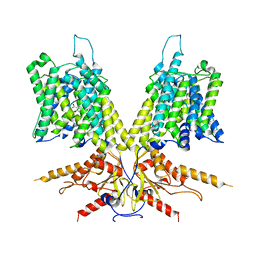 | | Cryo-EM Structure of dolphin Prestin: Inhibited I (Chloride + Salicylate) | | Descriptor: | 2-HYDROXYBENZOIC ACID, Prestin | | Authors: | Bavi, N, Clark, M.D, Contreras, G.F, Shen, R, Reddy, B.G, Milewski, W, Perozo, E. | | Deposit date: | 2021-09-20 | | Release date: | 2021-11-03 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The conformational cycle of prestin underlies outer-hair cell electromotility.
Nature, 600, 2021
|
|
7S9B
 
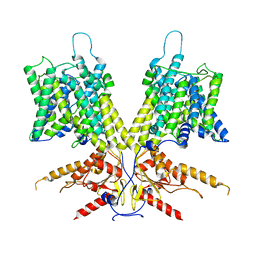 | | Cryo-EM Structure of dolphin Prestin: Sensor Down I (Expanded) state | | Descriptor: | Prestin | | Authors: | Bavi, N, Clark, M.D, Contreras, G.F, Shen, R, Reddy, B.G, Milewski, W, Perozo, E. | | Deposit date: | 2021-09-20 | | Release date: | 2021-11-03 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The conformational cycle of prestin underlies outer-hair cell electromotility.
Nature, 600, 2021
|
|
5UJ2
 
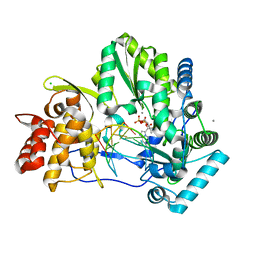 | | Crystal structure of HCV NS5B genotype 2A JFH-1 isolate with S15G E86Q E87Q C223H V321I mutations and Delta8 neta hairpoin loop deletion in complex with GS-639476 (diphsohate version of GS-9813), Mn2+ and symmetrical primer template 5'-AUAAAUUU | | Descriptor: | (1S)-1-(4-aminoimidazo[2,1-f][1,2,4]triazin-7-yl)-1,4-anhydro-2-deoxy-2-fluoro-5-O-[(S)-hydroxy(phosphonooxy)phosphoryl]-2-methyl-D-ribitol, CHLORIDE ION, Genome polyprotein, ... | | Authors: | Edwards, T.E, Fox III, D, Appleby, T.C, Murakami, E, Rey, A, McGrath, M.E. | | Deposit date: | 2017-01-16 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of a 2'-fluoro-2'-C-methyl C-nucleotide HCV polymerase inhibitor and a phosphoramidate prodrug with favorable properties.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
1NME
 
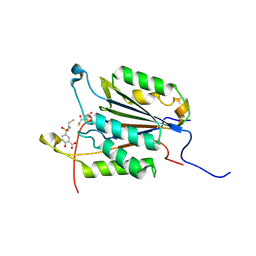 | | Structure of Casp-3 with tethered salicylate | | Descriptor: | 2-HYDROXY-5-(2-MERCAPTO-ETHYLSULFAMOYL)-BENZOIC ACID, 3-(2-MERCAPTO-ACETYLAMINO)-4-OXO-PENTANOIC ACID, Caspase-3 | | Authors: | Erlanson, D.A, Lam, J, Wiesmann, C, Luong, T.N, Simmons, B, DeLano, W, Choong, I.C, Flanagan, M, Lee, D, O'Brian, T. | | Deposit date: | 2003-01-09 | | Release date: | 2003-03-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | In situ assembly of enzyme inhibitors using extended tethering.
Nat.Biotechnol., 21, 2003
|
|
1NMQ
 
 | | Extendend Tethering: In Situ Assembly of Inhibitors | | Descriptor: | 3-(3-{2-[(5-METHANESULFONYL-THIOPHENE-2-CARBONYL)-AMINO]-ETHYLDISULFANYLMETHYL}- BENZENESULFONYLAMINO)-4-OXO-PENTANOIC ACID, Caspase-3 | | Authors: | Erlanson, D.A, Lam, J, Wiesmann, C, Luong, T.N, Simmons, R.L, DeLano, W, Choong, I.C, Flanagan, M, Lee, D, O'Brian, T. | | Deposit date: | 2003-01-10 | | Release date: | 2003-03-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | In situ assembly of enzyme inhibitors using extended tethering.
Nat.Biotechnol., 21, 2003
|
|
3D14
 
 | | Crystal structure of mouse Aurora A (Asn186->Gly, Lys240->Arg, Met302->Leu) in complex with 1-{5-[2-(thieno[3,2-d]pyrimidin-4-ylamino)-ethyl]- thiazol-2-yl}-3-(3-trifluoromethyl-phenyl)-urea | | Descriptor: | 1-{5-[2-(thieno[3,2-d]pyrimidin-4-ylamino)ethyl]-1,3-thiazol-2-yl}-3-[3-(trifluoromethyl)phenyl]urea, serine/threonine kinase 6 | | Authors: | Elling, R.A, Baskaran, S, Allen, D.A, Oslob, J.D, Romanowski, M.J. | | Deposit date: | 2008-05-04 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a potent and selective aurora kinase inhibitor.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3DBE
 
 | |
3DBD
 
 | |
3D15
 
 | | Crystal structure of mouse Aurora A (Asn186->Gly, Lys240->Arg, Met302->Leu) in complex with 1-(3-chloro-phenyl)-3-{5-[2-(thieno[3,2-d]pyrimidin-4-ylamino)- ethyl]-thiazol-2-yl}-urea [SNS-314] | | Descriptor: | 1-(3-chlorophenyl)-3-{5-[2-(thieno[3,2-d]pyrimidin-4-ylamino)ethyl]-1,3-thiazol-2-yl}urea, serine/threonine kinase 6 | | Authors: | Elling, R.A, Bui, M, Allen, D.A, Oslob, J.D, Romanowski, M.J. | | Deposit date: | 2008-05-04 | | Release date: | 2009-05-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a potent and selective aurora kinase inhibitor.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3DBC
 
 | | Crystal structure of an activated (Thr->Asp) Polo-like kinase 1 (Plk1) catalytic domain in complex with Compound 257 | | Descriptor: | 3-[3-(3-methyl-6-{[(1S)-1-phenylethyl]amino}-1H-pyrazolo[4,3-c]pyridin-1-yl)phenyl]propanamide, Polo-like kinase 1 | | Authors: | Elling, R.A, Zhu, J, Barr, K.J, Romanowski, M.J. | | Deposit date: | 2008-05-31 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Design and synthesis of 2-amino-pyrazolopyridines as Polo-like kinase 1 inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3DBF
 
 | | Crystal structure of an activated (Thr->Asp) Polo-like kinase 1 (Plk1) catalytic domain in complex with Compound 562 | | Descriptor: | 4-({1-[3-(3-amino-3-oxopropyl)-5-chlorophenyl]-3-methyl-1H-pyrazolo[4,3-c]pyridin-6-yl}amino)-3-methoxy-N-(1-methylpipe ridin-4-yl)benzamide, Polo-like kinase | | Authors: | Elling, R.A, Zhu, J, Barr, K.J, Romanowski, M.J. | | Deposit date: | 2008-05-31 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Design and synthesis of 2-amino-pyrazolopyridines as Polo-like kinase 1 inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1NMS
 
 | | Caspase-3 tethered to irreversible inhibitor | | Descriptor: | 5-[4-(1-CARBOXYMETHYL-2-OXO-PROPYLCARBAMOYL)-BENZYLSULFAMOYL]-2-HYDROXY-BENZOIC ACID, Caspase-3 | | Authors: | Erlanson, D.A, Lam, J, Wiesmann, C, Luong, T.N, Simmons, R.L, DeLano, W.L, Choong, I.C, Flanagan, W.M, Lee, D, O'Brian, T. | | Deposit date: | 2003-01-10 | | Release date: | 2003-03-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In situ assembly of enzyme inhibitors using extended tethering.
Nat.Biotechnol., 21, 2003
|
|
2XX2
 
 | | Macrolactone Inhibitor bound to HSP90 N-term | | Descriptor: | (5E)-13-CHLORO-14,16-DIHYDROXY-3,4,7,8,9,10-HEXAHYDRO-2-BENZAZACYCLOTETRADECINE-1,11(2H,12H)-DIONE, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82, GLYCEROL | | Authors: | Moody, C.J, Prodromou, C, Pearl, L.H, Roe, S.M. | | Deposit date: | 2010-11-08 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Targeting the Hsp90 Molecular Chaperone with Novel Macrolactams. Synthesis, Structural, Binding, and Cellular Studies.
Acs Chem.Biol., 6, 2011
|
|
4OGA
 
 | |
1G6T
 
 | | STRUCTURE OF EPSP SYNTHASE LIGANDED WITH SHIKIMATE-3-PHOSPHATE | | Descriptor: | EPSP SYNTHASE, FORMIC ACID, PHOSPHATE ION, ... | | Authors: | Schonbrunn, E, Eschenburg, S, Shuttleworth, W, Schloss, J.V, Amrhein, N, Evans, J.N.S, Kabsch, W. | | Deposit date: | 2000-11-07 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1G6S
 
 | | STRUCTURE OF EPSP SYNTHASE LIGANDED WITH SHIKIMATE-3-PHOSPHATE AND GLYPHOSATE | | Descriptor: | EPSP SYNTHASE, FORMIC ACID, GLYPHOSATE, ... | | Authors: | Schonbrunn, E, Eschenburg, S, Shuttleworth, W, Schloss, J.V, Amrhein, N, Evans, J.N.S, Kabsch, W. | | Deposit date: | 2000-11-07 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2XX4
 
 | | Macrolactone Inhibitor bound to HSP90 N-term | | Descriptor: | (E)-ETHYL 13-CHLORO-14,16-DIHYDROXY-1,11-DIOXO-1,2,3,4,7,8,9,10,11,12-DECAHYDROBENZO[C][1]AZACYCLOTETRADECINE-10-CARBOXYLATE, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82 | | Authors: | Moody, C.J, Prodromou, C, Pearl, L.H, Roe, S.M. | | Deposit date: | 2010-11-08 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Targeting the Hsp90 Molecular Chaperone with Novel Macrolactams. Synthesis, Structural, Binding, and Cellular Studies.
Acs Chem.Biol., 6, 2011
|
|
2XX5
 
 | | Macrolactone Inhibitor bound to HSP90 N-term | | Descriptor: | (5E,10R)-N-BENZYL-13-CHLORO-14,16-DIHYDROXY-1,11-DIOXO-1,2,3,4,7,8,9,10,11,12-DECAHYDRO-2-BENZAZACYCLOTETRADECINE-10-CARBOXAMIDE, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82, GLYCEROL | | Authors: | Moody, C.J, Prodromou, C, Pearl, L.H, Roe, S.M. | | Deposit date: | 2010-11-08 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Targeting the Hsp90 Molecular Chaperone with Novel Macrolactams. Synthesis, Structural, Binding, and Cellular Studies.
Acs Chem.Biol., 6, 2011
|
|
2XD6
 
 | | Hsp90 complexed with a resorcylic acid macrolactone. | | Descriptor: | (5Z)-13-CHLORO-14,16-DIHYDROXY-1,11-DIOXO-3,4,7,8,9,10,11,12-OCTAHYDRO-1H-2-BENZOXACYCLOTETRADECINE-6-CARBALDEHYDE, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82, GLYCEROL | | Authors: | Roe, S.M, Prodromou, C, Pearl, L.H, Moody, C.J. | | Deposit date: | 2010-04-29 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibition of Hsp90 with Resorcylic Acid Macrolactones. Synthesis and Binding Studies.
Chemistry, 16, 2010
|
|
4ASB
 
 | | The structure of modified benzoquinone ansamycins bound to yeast N- terminal Hsp90 | | Descriptor: | (4E,6E,8S,9R,10E,12R,13R,14S,16R)-19-{[2-(dimethylamino)ethyl]amino}-13-hydroxy-8,14-dimethoxy-4,10,12,16,21-pentamethyl-3,20,22-trioxo-2-azabicyclo[16.3.1]docosa-1(21),4,6,10,18-pentaen-9-yl carbamate, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82 | | Authors: | Roe, S.M, Prodromou, C. | | Deposit date: | 2012-04-30 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Synthesis of 19-substituted geldanamycins with altered conformations and their binding to heat shock protein Hsp90.
Nat Chem, 5, 2013
|
|
