2QFR
 
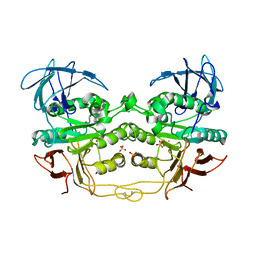 | | Crystal structure of red kidney bean purple acid phosphatase with bound sulfate | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, FE (III) ION, ... | | Authors: | Guddat, L.W, Schenk, G, Gahan, L.R, Elliot, T.W, Leung, E. | | Deposit date: | 2007-06-27 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of a purple acid phosphatase, representing different steps of this enzyme's catalytic cycle.
Bmc Struct.Biol., 8, 2008
|
|
5DKP
 
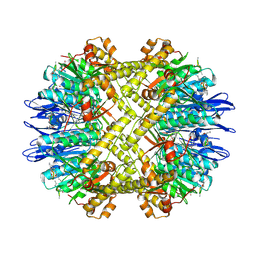 | | Crystal Structure of N. meningitidis ClpP in complex with agonist ADEP A54556. | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, POTASSIUM ION, SODIUM ION, ... | | Authors: | Goodreid, J.D, Janetzko, J, Santa Maria Jr, J.P, Wong, K, Leung, E, Eger, B.T, Bryson, S, Pai, E.F, Gray-Owen, S.D, Walker, S, Houry, W.A, Batey, R.A. | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.381 Å) | | Cite: | Development and Characterization of Potent Cyclic Acyldepsipeptide Analogues with Increased Antimicrobial Activity.
J.Med.Chem., 59, 2016
|
|
2QFP
 
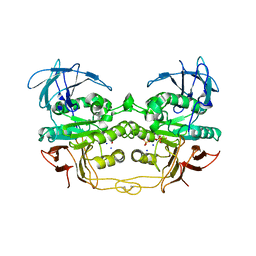 | | Crystal structure of red kidney bean purple acid phosphatase in complex with fluoride | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FE (III) ION, FLUORIDE ION, ... | | Authors: | Guddat, L.W, Schenk, G.S, Gahan, L.R, Elliot, T.W, Leung, E. | | Deposit date: | 2007-06-27 | | Release date: | 2008-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of a purple acid phosphatase, representing different steps of this enzyme's catalytic cycle.
Bmc Struct.Biol., 8, 2008
|
|
2XQW
 
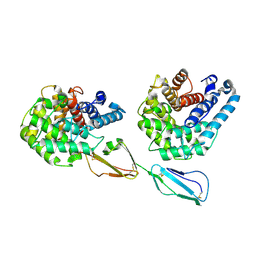 | | Structure of Factor H domains 19-20 in complex with complement C3d | | Descriptor: | COMPLEMENT C3, COMPLEMENT FACTOR H | | Authors: | Kajander, T, Lehtinen, M.J, Hyvarinen, S, Bhattacharjee, A, Leung, E, Isenman, D.E, Meri, S, Jokiranta, T.S, Goldman, A. | | Deposit date: | 2010-09-07 | | Release date: | 2011-02-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.306 Å) | | Cite: | Dual Interaction of Factor H with C3D and Glycosaminoglycans in Host-Nonhost Discrimination by Complement.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3HLN
 
 | | Crystal structure of ClpP A153C mutant with inter-heptamer disulfide bonds | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, CALCIUM ION | | Authors: | Kimber, M.S, Yu, A.Y.H, Borg, M, Chan, H.S, Houry, W.A. | | Deposit date: | 2009-05-27 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and Theoretical Studies Indicate that the Cylindrical Protease ClpP Samples Extended and Compact Conformations.
Structure, 18, 2010
|
|
6W9T
 
 | |
2JVG
 
 | | Structure of C3-binding domain 4 of Staphylococcus aureus protein Sbi | | Descriptor: | IgG-binding protein SBI | | Authors: | Upadhyay, A, Burman, J, Clark, E.A, van den Elsen, J.M.H, Bagby, S. | | Deposit date: | 2007-09-20 | | Release date: | 2008-06-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of the C3 binding region of Staphylococcus aureus immune subversion protein Sbi.
J.Biol.Chem., 283, 2008
|
|
2JVH
 
 | | Structure of C3-binding domain 4 of S. aureus protein Sbi | | Descriptor: | IgG-binding protein SBI | | Authors: | Upadhyay, A, Burman, J, Clark, E.A, van den Elsen, J.M.H, Bagby, S. | | Deposit date: | 2007-09-20 | | Release date: | 2008-06-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of the C3 binding region of Staphylococcus aureus immune subversion protein Sbi.
J.Biol.Chem., 283, 2008
|
|
7UVR
 
 | | Crystal structure of human ClpP protease in complex with TR-65 | | Descriptor: | 3-{[(10R)-4-[(4-chlorophenyl)methyl]-5-oxo-1,2,4,5,8,9-hexahydroimidazo[1,2-a]pyrido[3,4-e]pyrimidin-7(6H)-yl]methyl}benzonitrile, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | Authors: | Mabanglo, M.F, Houry, W.A. | | Deposit date: | 2022-05-02 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Potent ClpP agonists with anticancer properties bind with improved structural complementarity and alter the mitochondrial N-terminome.
Structure, 31, 2023
|
|
7UVM
 
 | | Crystal structure of human ClpP protease in complex with TR-27 | | Descriptor: | (10R)-4-[(4-chlorophenyl)methyl]-7-[(3-ethynylphenyl)methyl]-2,4,6,7,8,9-hexahydroimidazo[1,2-a]pyrido[3,4-e]pyrimidin-5(1H)-one, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | Authors: | Mabanglo, M.F, Houry, W.A. | | Deposit date: | 2022-05-02 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Potent ClpP agonists with anticancer properties bind with improved structural complementarity and alter the mitochondrial N-terminome.
Structure, 31, 2023
|
|
7UVN
 
 | | Crystal structure of human ClpP protease in complex with TR-57 | | Descriptor: | 3-({3-[(4-chlorophenyl)methyl]-1-methyl-2,4-dioxo-1,3,4,5,7,8-hexahydropyrido[4,3-d]pyrimidin-6(2H)-yl}methyl)benzonitrile, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | Authors: | Mabanglo, M.F, Houry, W.A. | | Deposit date: | 2022-05-02 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Potent ClpP agonists with anticancer properties bind with improved structural complementarity and alter the mitochondrial N-terminome.
Structure, 31, 2023
|
|
7UVU
 
 | | Crystal structure of human ClpP protease in complex with TR-107 | | Descriptor: | 3-({3-[(4-chlorophenyl)methyl]-4-oxo-3,5,7,8-tetrahydropyrido[4,3-d]pyrimidin-6(4H)-yl}methyl)benzonitrile, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | Authors: | Mabanglo, M.F, Houry, W.A. | | Deposit date: | 2022-05-02 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Potent ClpP agonists with anticancer properties bind with improved structural complementarity and alter the mitochondrial N-terminome.
Structure, 31, 2023
|
|
7UW0
 
 | | Crystal structure of human ClpP protease in complex with TR-133 | | Descriptor: | 3-({3-[(4-bromophenyl)methyl]-4-oxo-3,5,7,8-tetrahydropyrido[4,3-d]pyrimidin-6(4H)-yl}methyl)benzonitrile, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | Authors: | Mabanglo, M.F, Houry, W.A. | | Deposit date: | 2022-05-02 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Potent ClpP agonists with anticancer properties bind with improved structural complementarity and alter the mitochondrial N-terminome.
Structure, 31, 2023
|
|
6NAW
 
 | |
6NAQ
 
 | | Crystal structure of Neisseria meningitidis ClpP protease in Apo form | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, POTASSIUM ION | | Authors: | Houry, W.A, Mabanglo, M.F, Pai, E.F, Eger, B.T, Bryson, S. | | Deposit date: | 2018-12-06 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.022 Å) | | Cite: | ClpP protease activation results from the reorganization of the electrostatic interaction networks at the entrance pores.
Commun Biol, 2, 2019
|
|
6NB1
 
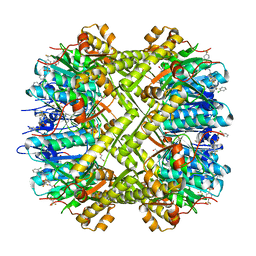 | | Crystal structure of Escherichia coli ClpP protease complexed with small molecule activator, ACP1-06 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, GLYCEROL, N-{2-[(2-chlorophenyl)sulfanyl]ethyl}-2-methyl-2-{[5-(trifluoromethyl)pyridin-2-yl]sulfonyl}propanamide | | Authors: | Mabanglo, M.F, Houry, W.A, Eger, B.T, Bryson, S, Pai, E.F. | | Deposit date: | 2018-12-06 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | ClpP protease activation results from the reorganization of the electrostatic interaction networks at the entrance pores.
Commun Biol, 2, 2019
|
|
6NAY
 
 | |
6NAH
 
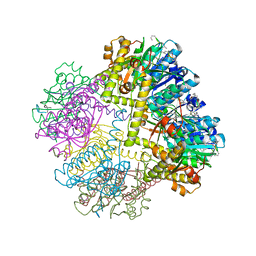 | |
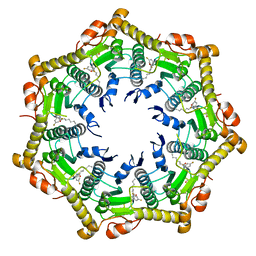
6BBA
 
 | |
