3SXO
 
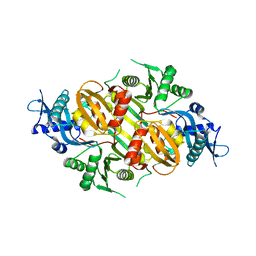 | | Mycobacterium tuberculosis Eis protein initiates modulation of host immune responses by acetylation of DUSP16/MKP-7 | | 分子名称: | Enhanced intracellular survival protein | | 著者 | Kim, K.H, An, D.R, Yoon, J.Y, Kim, H.S, Yoon, H.J, Song, J.S, Im, H.N, Kim, J, Kim, D.J, Lee, S.J, Kim, H.J, Lee, J.Y, Suh, S.W. | | 登録日 | 2011-07-15 | | 公開日 | 2012-07-25 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Mycobacterium tuberculosis Eis protein initiates modulation of host immune responses by acetylation of DUSP16/MKP-7
To be Published
|
|
7D04
 
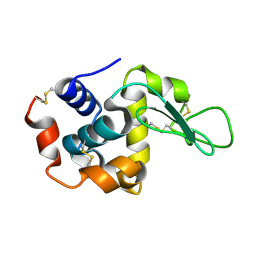 | |
7D05
 
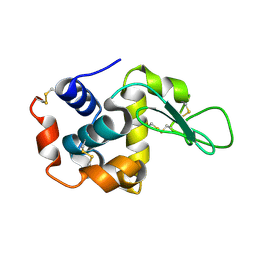 | |
7D01
 
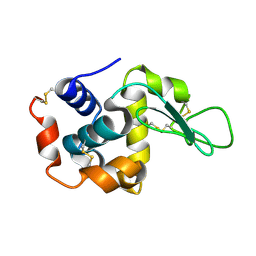 | |
7D02
 
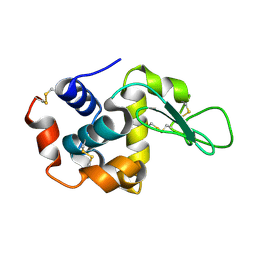 | |
3VPS
 
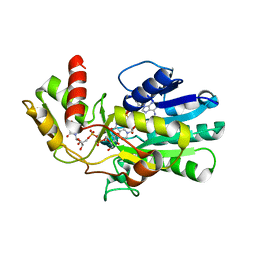 | | Structure of a novel NAD dependent-NDP-hexosamine 5,6-dehydratase, TunA, involved in tunicamycin biosynthesis | | 分子名称: | NAD-dependent epimerase/dehydratase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | 著者 | Wyszynski, F.J, Lee, S.S, Yabe, T, Wang, H, Gomez-Escribano, J.P, Bibb, M.J, Lee, S.J, Davies, G.J, Davis, B.G. | | 登録日 | 2012-03-12 | | 公開日 | 2012-04-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Biosynthesis of nucleoside antibiotic tunicamycin proceeds via unique exo-glycal intermediates
To be published
|
|
3QY8
 
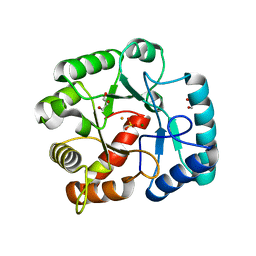 | | Crystal structures of YwqE from Bacillus subtilis and CpsB from Streptococcus pneumoniae, unique metal-dependent tyrosine phosphatases | | 分子名称: | FE (III) ION, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Kim, H.S, Lee, S.J, Yoon, H.J, An, D.R, Kim, D.J, Kim, S.-J, Suh, S.W. | | 登録日 | 2011-03-03 | | 公開日 | 2011-06-08 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structures of YwqE from Bacillus subtilis and CpsB from Streptococcus pneumoniae, unique metal-dependent tyrosine phosphatases.
J.Struct.Biol., 175, 2011
|
|
3QY7
 
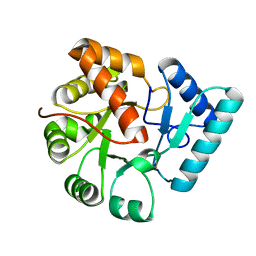 | | Crystal structures of YwqE from Bacillus subtilis and CpsB from Streptococcus pneumoniae, unique metal-dependent tyrosine phosphatases | | 分子名称: | FE (III) ION, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Kim, H.S, Lee, S.J, Yoon, H.J, An, D.R, Kim, D.J, Kim, S.-J, Suh, S.W. | | 登録日 | 2011-03-03 | | 公開日 | 2011-06-08 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Crystal structures of YwqE from Bacillus subtilis and CpsB from Streptococcus pneumoniae, unique metal-dependent tyrosine phosphatases.
J.Struct.Biol., 175, 2011
|
|
3QY6
 
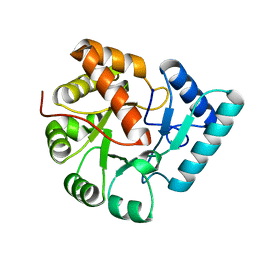 | | Crystal structures of YwqE from Bacillus subtilis and CpsB from Streptococcus pneumoniae, unique metal-dependent tyrosine phosphatases | | 分子名称: | FE (III) ION, MAGNESIUM ION, Tyrosine-protein phosphatase YwqE | | 著者 | Kim, H.S, Lee, S.J, Yoon, H.J, An, D.R, Kim, D.J, Kim, S.-J, Suh, S.W. | | 登録日 | 2011-03-03 | | 公開日 | 2011-06-08 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structures of YwqE from Bacillus subtilis and CpsB from Streptococcus pneumoniae, unique metal-dependent tyrosine phosphatases.
J.Struct.Biol., 175, 2011
|
|
8E2R
 
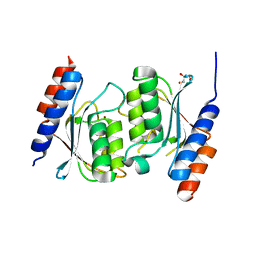 | |
8E2S
 
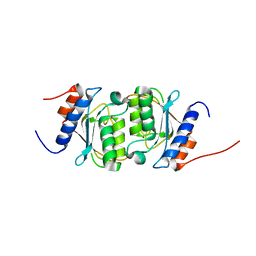 | |
8E2P
 
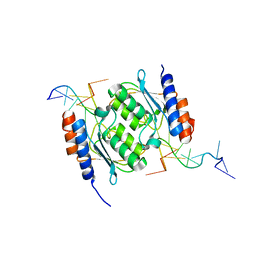 | |
8E2Q
 
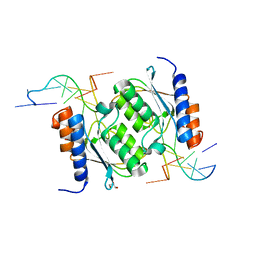 | | Crystal structure of TadAC-1.17 in a complex with ssDNA | | 分子名称: | DNA (5'-D(P*GP*CP*GP*GP*CP*TP*(D8A)P*CP*GP*GP*A)-3'), GLYCEROL, ZINC ION, ... | | 著者 | Feliciano, P.R, Lee, S.J, Ciaramella, G. | | 登録日 | 2022-08-15 | | 公開日 | 2023-01-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Improved cytosine base editors generated from TadA variants.
Nat.Biotechnol., 41, 2023
|
|
6L2U
 
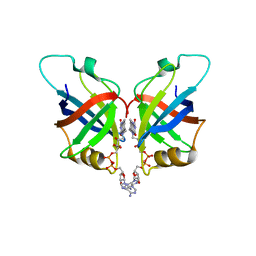 | | Soluble methane monooxygenase reductase FAD-binding domain from Methylosinus sporium. | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Methane monooxygenase | | 著者 | Park, J.H, Ha, S.C, Rao, Z, Yoo, H, Yoon, C, Kim, S.Y, Kim, D.S, Lee, S.J. | | 登録日 | 2019-10-07 | | 公開日 | 2021-03-03 | | 最終更新日 | 2021-12-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Elucidation of the electron transfer environment in the MMOR FAD-binding domain from Methylosinus sporium 5.
Dalton Trans, 50, 2021
|
|
5GKV
 
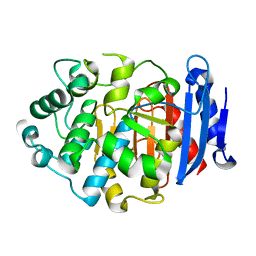 | | Crystal Structure of a Novel Penicillin-Binding Protein (PBP) Homolog from Caulobacter crescentus | | 分子名称: | Esterase A | | 著者 | Ngo, T.D, Ryu, B.H, Kim, B.Y, Yoo, W.K, Lee, E.J, Lee, S.J, Kim, T.D, Kim, K.K. | | 登録日 | 2016-07-07 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | Biochemical and Structural Analysis of a Novel Penicillin-Binding Protein (PBP) Homolog from Caulobacter crescentus
To Be Published
|
|
5YGM
 
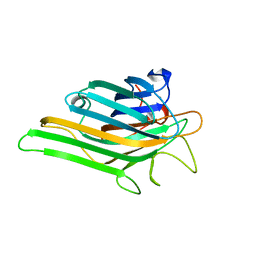 | |
6AHG
 
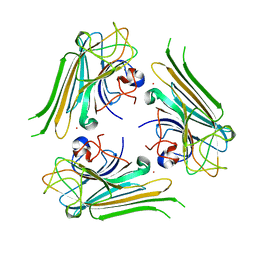 | | Trimeric structure of concanavalin A from Canavalia ensiformis | | 分子名称: | CADMIUM ION, CALCIUM ION, Concanavalin-A,Concanavalin-A | | 著者 | Park, J.H, Kim, D.S, Park, Y.R, Lee, S.J. | | 登録日 | 2018-08-18 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.83 Å) | | 主引用文献 | Cadmium-substituted concanavalin A and its trimeric complexation
J. Microbiol. Biotechnol., 28(12), 2018
|
|
3DCM
 
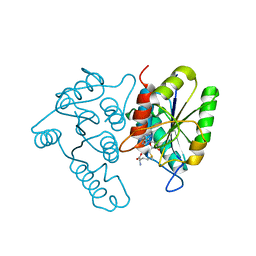 | | Crystal structure of the Thermotoga maritima SPOUT family RNA-methyltransferase protein Tm1570 in complex with S-adenosyl-L-methionine | | 分子名称: | S-ADENOSYLMETHIONINE, Uncharacterized protein TM_1570 | | 著者 | Kim, D.J, Kim, H.S, Lee, S.J, Suh, S.W. | | 登録日 | 2008-06-04 | | 公開日 | 2008-12-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of Thermotoga maritima SPOUT superfamily RNA methyltransferase Tm1570 in complex with S-adenosyl-L-methionine
Proteins, 74, 2009
|
|
3GDE
 
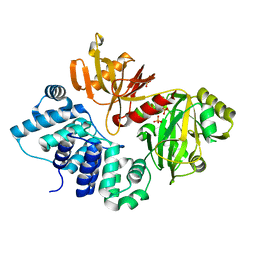 | | The closed conformation of ATP-dependent DNA ligase from Archaeoglobus fulgidus | | 分子名称: | DNA ligase, PHOSPHATE ION | | 著者 | Kim, D.J, Kim, H.-W, Kim, O, Kim, H.S, Lee, S.J, Suh, S.W. | | 登録日 | 2009-02-24 | | 公開日 | 2009-12-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | ATP-dependent DNA ligase from Archaeoglobus fulgidus displays a tightly closed conformation
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3A6P
 
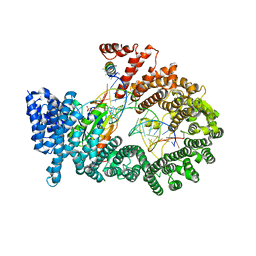 | | Crystal structure of Exportin-5:RanGTP:pre-miRNA complex | | 分子名称: | 13-mer peptide, Exportin-5, GTP-binding nuclear protein Ran, ... | | 著者 | Okada, C, Yamashita, E, Lee, S.J, Shibata, S, Katahira, J, Nakagawa, A, Yoneda, Y, Tsukihara, T. | | 登録日 | 2009-09-07 | | 公開日 | 2009-12-08 | | 最終更新日 | 2012-04-25 | | 実験手法 | X-RAY DIFFRACTION (2.92 Å) | | 主引用文献 | A high-resolution structure of the pre-microRNA nuclear export machinery
Science, 326, 2009
|
|
3B1F
 
 | | Crystal structure of prephenate dehydrogenase from Streptococcus mutans | | 分子名称: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative prephenate dehydrogenase | | 著者 | Ku, H.K, Do, N.H, Song, J.S, Choi, S, Shin, M.H, Kim, K.J, Lee, S.J. | | 登録日 | 2011-07-02 | | 公開日 | 2011-10-26 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of prephenate dehydrogenase from Streptococcus mutans.
Int.J.Biol.Macromol., 49, 2011
|
|
2F5T
 
 | | Crystal Structure of the sugar binding domain of the archaeal transcriptional regulator TrmB | | 分子名称: | IMIDAZOLE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, archaeal transcriptional regulator TrmB | | 著者 | Krug, M, Lee, S.J, Diederichs, K, Boos, W, Welte, W. | | 登録日 | 2005-11-27 | | 公開日 | 2006-02-21 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Crystal Structure of the Sugar Binding Domain of the Archaeal Transcriptional Regulator TrmB
J.Biol.Chem., 281, 2006
|
|
1UMI
 
 | | Structural basis of sugar-recognizing ubiquitin ligase | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, F-box only protein 2 | | 著者 | Mizushima, T, Hirao, T, Yoshida, Y, Lee, S.J, Chiba, T, Iwai, K, Yamaguchi, Y, Kato, K, Tsukihara, T, Tanaka, K. | | 登録日 | 2003-10-01 | | 公開日 | 2004-04-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis of sugar-recognizing ubiquitin ligase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1UMH
 
 | | Structural basis of sugar-recognizing ubiquitin ligase | | 分子名称: | F-box only protein 2, NICKEL (II) ION | | 著者 | Mizushima, T, Hirao, T, Yoshida, Y, Lee, S.J, Chiba, T, Iwai, K, Yamaguchi, Y, Kato, K, Tsukihara, T, Tanaka, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-10-01 | | 公開日 | 2004-04-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis of sugar-recognizing ubiquitin ligase
NAT.STRUCT.MOL.BIOL., 11, 2004
|
|
1V8Z
 
 | | X-ray crystal structure of the Tryptophan Synthase b2 Subunit from Hyperthermophile, Pyrococcus furiosus | | 分子名称: | PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, Tryptophan synthase beta chain 1 | | 著者 | Hioki, Y, Ogasahara, K, Lee, S.J, Ma, J, Ishida, M, Yamagata, Y, Matsuura, Y, Ota, M, Kuramitsu, S, Yutani, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2004-01-15 | | 公開日 | 2005-02-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | The crystal structure of the tryptophan synthase beta subunit from the hyperthermophile Pyrococcus furiosus. Investigation of stabilization factors
Eur.J.Biochem., 271, 2004
|
|
