2WWX
 
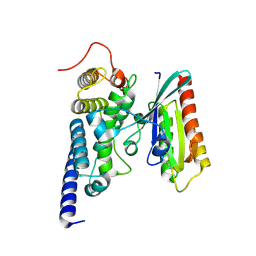 | | Crystal structure of the SidM/DrrA(GEF/GDF domain)-Rab1(GTPase domain) complex | | Descriptor: | DRRA, RAS-RELATED PROTEIN RAB-1 | | Authors: | Suh, H.Y, Lee, D.W, Woo, J.S, Oh, B.H. | | Deposit date: | 2009-10-30 | | Release date: | 2009-12-08 | | Last modified: | 2015-04-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insights Into the Dual Nucleotide Exchange and Gdi Displacement Activity of Sidm/Drra
Embo J., 29, 2010
|
|
4YVM
 
 | |
5J4G
 
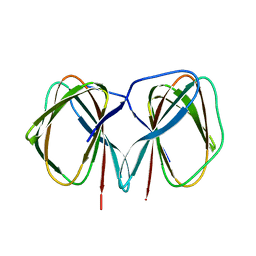 | | Crystal structure of the C-terminally His6-tagged HP0902, an uncharacterized protein from Helicobacter pylori 26695 | | Descriptor: | Uncharacterized protein | | Authors: | Sim, D.W, Lee, W.C, Kim, H.Y, Kim, J.H, Won, H.S. | | Deposit date: | 2016-04-01 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural identification of the lipopolysaccharide-binding capability of a cupin-family protein from Helicobacter pylori
FEBS Lett., 590, 2016
|
|
4DT3
 
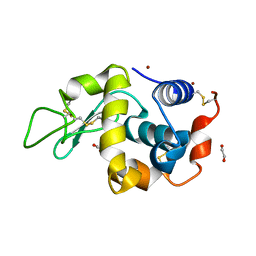 | | Crystal structure of zinc-charged lysozyme | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | An, Y.J, Jeong, C.S, Cha, S.S. | | Deposit date: | 2012-02-20 | | Release date: | 2012-09-12 | | Last modified: | 2013-07-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental phasing using zinc anomalous scattering
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5J4F
 
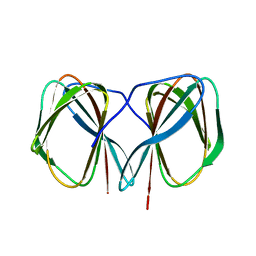 | | Crystal structure of the N-terminally His6-tagged HP0902, an uncharacterized protein from Helicobacter pylori 26695 | | Descriptor: | Uncharacterized protein | | Authors: | Sim, D.-W, Lee, W.-C, Kim, H.Y, Kim, J.-H, Won, H.-S. | | Deposit date: | 2016-04-01 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural identification of the lipopolysaccharide-binding capability of a cupin-family protein from Helicobacter pylori
FEBS Lett., 590, 2016
|
|
8ENJ
 
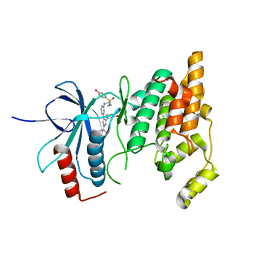 | |
3MVE
 
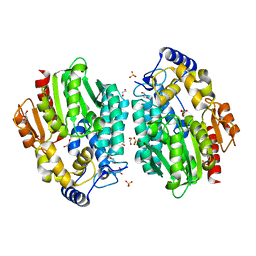 | | Crystal structure of a novel pyruvate decarboxylase | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, UPF0255 protein VV1_0328 | | Authors: | Cha, S.S, Jeong, C.S, An, Y.J. | | Deposit date: | 2010-05-04 | | Release date: | 2011-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | FrsA functions as a cofactor-independent decarboxylase to control metabolic flux.
Nat.Chem.Biol., 7, 2011
|
|
3OUR
 
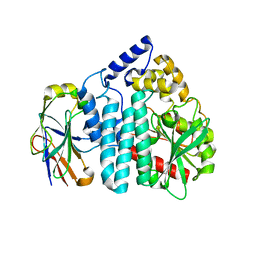 | |
2CK0
 
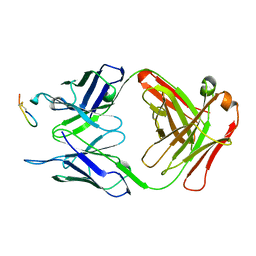 | |
5SV6
 
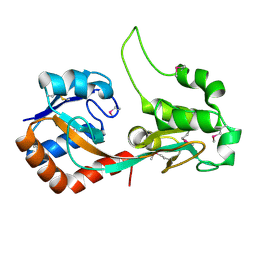 | |
1HA0
 
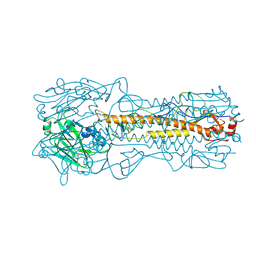 | | HEMAGGLUTININ PRECURSOR HA0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (HEMAGGLUTININ PRECURSOR), alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, J, Ho Lee, K, Steinhauer, D.A, Stevens, D.J, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1998-10-08 | | Release date: | 1998-10-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation.
Cell(Cambridge,Mass.), 95, 1998
|
|
5DBT
 
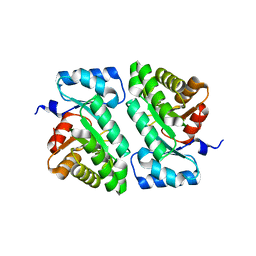 | |
5DBU
 
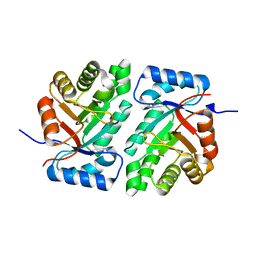 | |
6XE9
 
 | | 10S myosin II (smooth muscle) | | Descriptor: | Myosin II heavy chain (smooth muscle), Myosin light chain 9, Myosin light chain smooth muscle isoform | | Authors: | Tiwari, P, Craig, R, Padron, R. | | Deposit date: | 2020-06-12 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of the inhibited (10S) form of myosin II.
Nature, 588, 2020
|
|
4OWI
 
 | | peptide structure | | Descriptor: | p53LZ2 | | Authors: | Lee, J.-H. | | Deposit date: | 2014-02-02 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.202 Å) | | Cite: | Protein grafting of p53TAD onto a leucine zipper scaffold generates a potent HDM dual inhibitor.
Nat Commun, 5, 2014
|
|
3EUJ
 
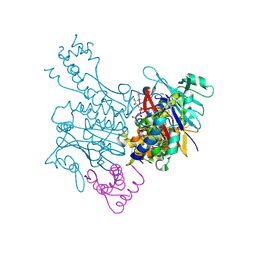 | | Crystal structure of MukE-MukF(residues 292-443)-MukB(head domain)-ATPgammaS complex, symmetric dimer | | Descriptor: | Chromosome partition protein mukB, Linker, Chromosome partition protein mukF, ... | | Authors: | Woo, J.S, Lim, J.H, Shin, H.C, Oh, B.H. | | Deposit date: | 2008-10-10 | | Release date: | 2009-01-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural studies of a bacterial condensin complex reveal ATP-dependent disruption of intersubunit interactions.
Cell(Cambridge,Mass.), 136, 2009
|
|
3EUK
 
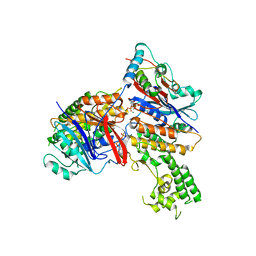 | | Crystal structure of MukE-MukF(residues 292-443)-MukB(head domain)-ATPgammaS complex, asymmetric dimer | | Descriptor: | Chromosome partition protein mukB, Linker, Chromosome partition protein mukE, ... | | Authors: | Woo, J.S, Lim, J.H, Shin, H.C, Oh, B.H. | | Deposit date: | 2008-10-10 | | Release date: | 2009-01-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural studies of a bacterial condensin complex reveal ATP-dependent disruption of intersubunit interactions.
Cell(Cambridge,Mass.), 136, 2009
|
|
3EUH
 
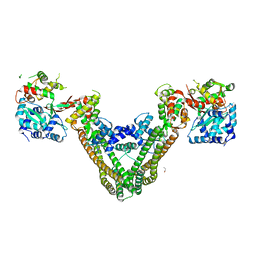 | | Crystal Structure of the MukE-MukF Complex | | Descriptor: | Chromosome partition protein mukF, GLYCINE, MukE | | Authors: | Suh, M.K, Ku, B, Ha, N.C, Woo, J.S, Oh, B.H. | | Deposit date: | 2008-10-10 | | Release date: | 2009-01-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of a bacterial condensin complex reveal ATP-dependent disruption of intersubunit interactions.
Cell(Cambridge,Mass.), 136, 2009
|
|
5EXJ
 
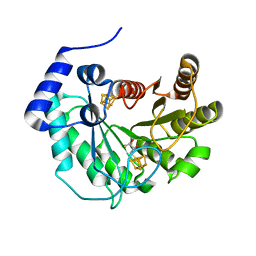 | | Crystal structure of M. tuberculosis lipoyl synthase at 1.64 A resolution | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, IRON/SULFUR CLUSTER, Lipoyl synthase | | Authors: | McLaughlin, M.I, Lanz, N.D, Goldman, P.J, Lee, K.-H, Booker, S.J, Drennan, C.L. | | Deposit date: | 2015-11-23 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystallographic snapshots of sulfur insertion by lipoyl synthase.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EXI
 
 | | Crystal structure of M. tuberculosis lipoyl synthase at 2.28 A resolution | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, IRON/SULFUR CLUSTER, Lipoyl synthase | | Authors: | McLaughlin, M.I, Lanz, N.D, Goldman, P.J, Lee, K.-H, Booker, S.J, Drennan, C.L. | | Deposit date: | 2015-11-23 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystallographic snapshots of sulfur insertion by lipoyl synthase.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EXK
 
 | | Crystal structure of M. tuberculosis lipoyl synthase with 6-thiooctanoyl peptide intermediate | | Descriptor: | 5'-DEOXYADENOSINE, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | McLaughlin, M.I, Lanz, N.D, Goldman, P.J, Lee, K.-H, Booker, S.J, Drennan, C.L. | | Deposit date: | 2015-11-23 | | Release date: | 2016-08-10 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystallographic snapshots of sulfur insertion by lipoyl synthase.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4ARZ
 
 | | The crystal structure of Gtr1p-Gtr2p complexed with GTP-GDP | | Descriptor: | GTP-BINDING PROTEIN GTR1, GTP-BINDING PROTEIN GTR2, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Jeong, J.H, Kim, Y.G. | | Deposit date: | 2012-04-27 | | Release date: | 2012-07-25 | | Last modified: | 2012-09-05 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of the Gtr1Pgtp-Gtr2Pgdp Complex Reveals Large Structural Rearrangements Triggered by GTP-to-Gdp Conversion
J.Biol.Chem., 287, 2012
|
|
7EYC
 
 | | Crystal structure of Tau and acetylated tau peptide antigen | | Descriptor: | ACETYLATED TAU PEPTIDE, antibody, Heavy chain, ... | | Authors: | Hong, M, Park, J. | | Deposit date: | 2021-05-30 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Monoclonal antibody Y01 prevents tauopathy progression induced by lysine 280-acetylated tau in cell and mouse models.
J.Clin.Invest., 133, 2023
|
|
1CK0
 
 | |
3CK0
 
 | |
