2EWT
 
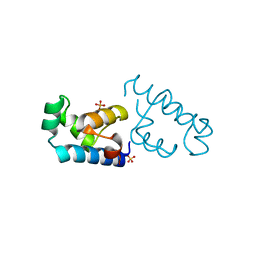 | | Crystal structure of the DNA-binding domain of BldD | | 分子名称: | SULFATE ION, putative DNA-binding protein | | 著者 | Kim, I.K, Lee, C.J, Kim, M.K, Kim, J.M, Kim, J.H, Yim, H.S, Cha, S.S, Kang, S.O. | | 登録日 | 2005-11-07 | | 公開日 | 2006-06-13 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Crystal structure of the DNA-binding domain of BldD, a central regulator of aerial mycelium formation in Streptomyces coelicolor A3(2)
Mol.Microbiol., 60, 2006
|
|
1WWL
 
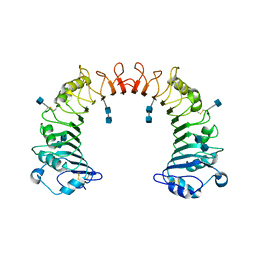 | | Crystal structure of CD14 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Monocyte differentiation antigen CD14 | | 著者 | Kim, J.-I, Lee, C.J, Jin, M.S, Lee, C.-H, Paik, S.-G, Lee, H, Lee, J.-O. | | 登録日 | 2005-01-06 | | 公開日 | 2005-02-22 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of CD14 and Its Implications for Lipopolysaccharide Signaling
J.Biol.Chem., 280, 2005
|
|
4FQG
 
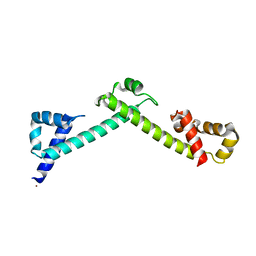 | | Crystal structure of the TCERG1 FF4-6 tandem repeat domain | | 分子名称: | CHLORIDE ION, NICKEL (II) ION, Transcription elongation regulator 1 | | 著者 | Liu, J, Fan, S, Lee, C.J, Greenleaf, A.L, Zhou, P. | | 登録日 | 2012-06-25 | | 公開日 | 2013-02-27 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Specific Interaction of the Transcription Elongation Regulator TCERG1 with RNA Polymerase II Requires Simultaneous Phosphorylation at Ser2, Ser5, and Ser7 within the Carboxyl-terminal Domain Repeat.
J.Biol.Chem., 288, 2013
|
|
3P3C
 
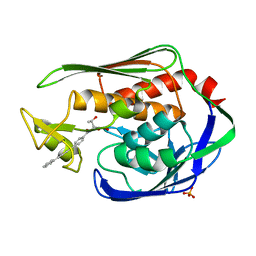 | | Crystal Structure of the Aquifex aeolicus LpxC/LPC-009 complex | | 分子名称: | N-[(1S,2R)-2-hydroxy-1-(hydroxycarbamoyl)propyl]-4-(4-phenylbuta-1,3-diyn-1-yl)benzamide, PHOSPHATE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | 著者 | Lee, C.-J, Zhou, P. | | 登録日 | 2010-10-04 | | 公開日 | 2011-01-05 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Species-specific and inhibitor-dependent conformations of LpxC: implications for antibiotic design.
Chem.Biol., 18, 2011
|
|
5DRP
 
 | | Structure of the AaLpxC/LPC-023 Complex | | 分子名称: | CHLORIDE ION, DIMETHYL SULFOXIDE, N~2~-{4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]benzoyl}-N-hydroxy-L-isoleucinamide, ... | | 著者 | Najeeb, J, Lee, C.-J, Zhou, P. | | 登録日 | 2015-09-16 | | 公開日 | 2016-03-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.889 Å) | | 主引用文献 | Drug design from the cryptic inhibitor envelope.
Nat Commun, 7, 2016
|
|
5DRR
 
 | | Crystal structure of the Pseudomonas aeruginosa LpxC/LPC-058 complex | | 分子名称: | 4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3S)-4,4-difluoro-3-hydroxy-1-(hydroxyamino)-3-methyl-1-oxobutan-2-yl]benzamide, NITRATE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | 著者 | Lee, C.-J, Najeeb, J, Zhou, P. | | 登録日 | 2015-09-16 | | 公開日 | 2016-03-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Drug design from the cryptic inhibitor envelope.
Nat Commun, 7, 2016
|
|
5DRQ
 
 | | Crystal structure of the Pseudomonas aeruginosa LpxC/LPC-040 complex | | 分子名称: | N-[(2S)-3-amino-1-(hydroxyamino)-3-methyl-1-oxobutan-2-yl]-4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]benzamide, NITRATE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | 著者 | Lee, C.-J, Najeeb, J, Zhou, P. | | 登録日 | 2015-09-16 | | 公開日 | 2016-03-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Drug design from the cryptic inhibitor envelope.
Nat Commun, 7, 2016
|
|
5DRO
 
 | | Structure of the Aquifex aeolicus LpxC/LPC-011 Complex | | 分子名称: | 4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3R)-3-hydroxy-1-(hydroxyamino)-1-oxobutan-2-yl]benzamide, ACETATE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Lee, C.-J, Najeeb, J, Zhou, P. | | 登録日 | 2015-09-16 | | 公開日 | 2016-03-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Drug design from the cryptic inhibitor envelope.
Nat Commun, 7, 2016
|
|
4IS9
 
 | | Crystal Structure of the Escherichia coli LpxC/L-161,240 complex | | 分子名称: | (4R)-2-(3,4-dimethoxy-5-propylphenyl)-N-hydroxy-4,5-dihydro-1,3-oxazole-4-carboxamide, ISOPROPYL ALCOHOL, SODIUM ION, ... | | 著者 | Lee, C.-J, Zhou, P. | | 登録日 | 2013-01-16 | | 公開日 | 2013-10-30 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Structural Basis of the Promiscuous Inhibitor Susceptibility of Escherichia coli LpxC.
Acs Chem.Biol., 9, 2014
|
|
4ISA
 
 | | Crystal Structure of the Escherichia coli LpxC/BB-78485 complex | | 分子名称: | (2R)-N-hydroxy-3-naphthalen-2-yl-2-[(naphthalen-2-ylsulfonyl)amino]propanamide, (4S,5S)-1,2-DITHIANE-4,5-DIOL, FORMIC ACID, ... | | 著者 | Lee, C.-J, Zhou, P. | | 登録日 | 2013-01-16 | | 公開日 | 2013-10-30 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural Basis of the Promiscuous Inhibitor Susceptibility of Escherichia coli LpxC.
Acs Chem.Biol., 9, 2014
|
|
3P3E
 
 | | Crystal Structure of the PSEUDOMONAS AERUGINOSA LpxC/LPC-009 complex | | 分子名称: | N-[(1S,2R)-2-hydroxy-1-(hydroxycarbamoyl)propyl]-4-(4-phenylbuta-1,3-diyn-1-yl)benzamide, NITRATE ION, SODIUM ION, ... | | 著者 | Lee, C.-J, Zhou, P. | | 登録日 | 2010-10-04 | | 公開日 | 2011-01-05 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | Species-specific and inhibitor-dependent conformations of LpxC: implications for antibiotic design.
Chem.Biol., 18, 2011
|
|
3P3G
 
 | | Crystal Structure of the Escherichia coli LpxC/LPC-009 complex | | 分子名称: | 4-ethynyl-N-[(1S,2R)-2-hydroxy-1-(oxocarbamoyl)propyl]benzamide, DIMETHYL SULFOXIDE, N-[(1S,2R)-2-hydroxy-1-(hydroxycarbamoyl)propyl]-4-(4-phenylbuta-1,3-diyn-1-yl)benzamide, ... | | 著者 | Lee, C.-J, Zhou, P. | | 登録日 | 2010-10-04 | | 公開日 | 2011-01-05 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Species-specific and inhibitor-dependent conformations of LpxC: implications for antibiotic design.
Chem.Biol., 18, 2011
|
|
4MQY
 
 | | Crystal Structure of the Escherichia coli LpxC/LPC-138 complex | | 分子名称: | 4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3R)-3-hydroxy-2-methyl-1-nitroso-1-oxobutan-2-yl]benzamide, 4-ethynyl-N-[(1S,2R)-2-hydroxy-1-(oxocarbamoyl)propyl]benzamide, DIMETHYL SULFOXIDE, ... | | 著者 | Lee, C.-J, Najeeb, J, Zhou, P. | | 登録日 | 2013-09-17 | | 公開日 | 2013-10-23 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.005 Å) | | 主引用文献 | Structural Basis of the Promiscuous Inhibitor Susceptibility of Escherichia coli LpxC.
Acs Chem.Biol., 9, 2014
|
|
2KRI
 
 | |
7KHM
 
 | |
5K8K
 
 | | Structure of the Haemophilus influenzae LpxH-lipid X complex | | 分子名称: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, ACETATE ION, GLYCEROL, ... | | 著者 | Cho, J, Lee, C.-J, Zhou, P. | | 登録日 | 2016-05-30 | | 公開日 | 2016-08-10 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structure of the essential Haemophilus influenzae UDP-diacylglucosamine pyrophosphohydrolase LpxH in lipid A biosynthesis.
Nat Microbiol, 1, 2016
|
|
8J26
 
 | | CryoEM structure of SARS CoV-2 RBD and Aptamer complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AM032-4, AM047-6, ... | | 著者 | Rahman, M.S, Jang, S.K, Lee, J.O. | | 登録日 | 2023-04-14 | | 公開日 | 2023-06-21 | | 最終更新日 | 2023-07-12 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure-Guided Development of Bivalent Aptamers Blocking SARS-CoV-2 Infection.
Molecules, 28, 2023
|
|
8J1Q
 
 | | CryoEM structure of SARS CoV-2 RBD and Aptamer complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AM032-0, AM047-0, ... | | 著者 | Rahman, M.S, Jang, S.K, Lee, J.O. | | 登録日 | 2023-04-13 | | 公開日 | 2023-06-21 | | 最終更新日 | 2023-07-12 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure-Guided Development of Bivalent Aptamers Blocking SARS-CoV-2 Infection.
Molecules, 28, 2023
|
|
5U86
 
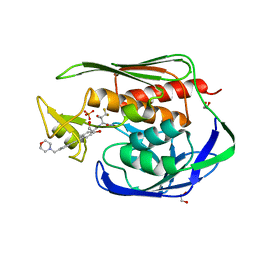 | | Structure of the Aquifex aeolicus LpxC/LPC-069 complex | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, N-[(2S,3S)-4,4-difluoro-3-hydroxy-1-(hydroxyamino)-3-methyl-1-oxobutan-2-yl]-4-({4-[(morpholin-4-yl)methyl]phenyl}ethynyl)benzamide, ... | | 著者 | Najeeb, J, Lee, C.-J, Zhou, P. | | 登録日 | 2016-12-13 | | 公開日 | 2017-08-09 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Curative Treatment of Severe Gram-Negative Bacterial Infections by a New Class of Antibiotics Targeting LpxC.
MBio, 8, 2017
|
|
3E7L
 
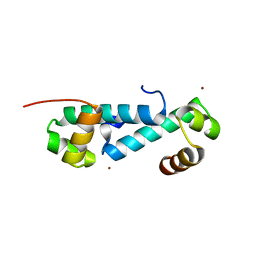 | | Crystal structure of sigma54 activator NtrC4's DNA binding domain | | 分子名称: | Transcriptional regulator (NtrC family), ZINC ION | | 著者 | Batchelor, J.D, Doucleff, M, Lee, C.-J, Matsubara, K, De Carlo, S, Heideker, J, Lamers, M.M, Pelton, J.G, Wemmer, D.E. | | 登録日 | 2008-08-18 | | 公開日 | 2008-11-25 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.252 Å) | | 主引用文献 | Structure and regulatory mechanism of Aquifex aeolicus NtrC4: variability and evolution in bacterial transcriptional regulation.
J.Mol.Biol., 384, 2008
|
|
3DZD
 
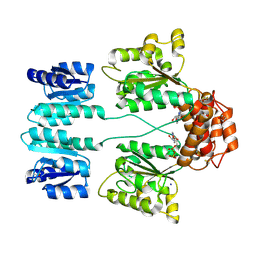 | | Crystal structure of sigma54 activator NTRC4 in the inactive state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, SODIUM ION, Transcriptional regulator (NtrC family) | | 著者 | Batchelor, J.D, Doucleff, M, Lee, C.-J, Matsubara, K, De Carlo, S, Heideker, J, Lamers, M.M, Pelton, J.G, Wemmer, D.E. | | 登録日 | 2008-07-29 | | 公開日 | 2008-11-25 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure and regulatory mechanism of Aquifex aeolicus NtrC4: variability and evolution in bacterial transcriptional regulation.
J.Mol.Biol., 384, 2008
|
|
1QPW
 
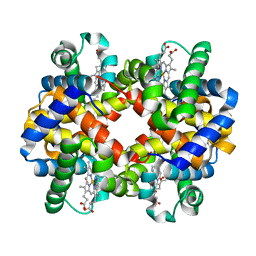 | | CRYSTAL STRUCTURE DETERMINATION OF PORCINE HEMOGLOBIN AT 1.8A RESOLUTION | | 分子名称: | OXYGEN MOLECULE, PORCINE HEMOGLOBIN (ALPHA SUBUNIT), PORCINE HEMOGLOBIN (BETA SUBUNIT), ... | | 著者 | Lu, T.-H, Panneerselvam, K, Liaw, Y.-C, Kan, P, Lee, C.-J. | | 登録日 | 1999-05-30 | | 公開日 | 1999-06-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure determination of porcine haemoglobin.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
4FJO
 
 | | Structure of the Rev1 CTD-Rev3/7-Pol kappa RIR complex | | 分子名称: | DNA polymerase kappa, DNA polymerase zeta catalytic subunit, DNA repair protein REV1, ... | | 著者 | Wojtaszek, J, Lee, C.-J, Zhou, P. | | 登録日 | 2012-06-11 | | 公開日 | 2012-08-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.718 Å) | | 主引用文献 | Structural basis of Rev1-mediated assembly of a quaternary vertebrate translesion polymerase complex consisting of Rev1, heterodimeric Pol zeta and Pol kappa
J.Biol.Chem., 287, 2012
|
|
3OP8
 
 | | Crystal structure of the domain V from beta2-glycoprotein I | | 分子名称: | Beta-2-glycoprotein 1, SULFATE ION | | 著者 | Kolyada, A, Lee, C.-J, De Biasio, A, Beglova, N. | | 登録日 | 2010-08-31 | | 公開日 | 2011-01-05 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A Novel Dimeric Inhibitor Targeting Beta2GPI in Beta2GPI/Antibody Complexes Implicated in Antiphospholipid Syndrome.
Plos One, 5, 2010
|
|
3PS1
 
 | | Crystal structure of the Escherichia Coli LPXC/LPC-011 complex | | 分子名称: | 4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3R)-3-hydroxy-1-(hydroxyamino)-1-oxobutan-2-yl]benzamide, 4-ethynyl-N-[(1S,2R)-2-hydroxy-1-(oxocarbamoyl)propyl]benzamide, DIMETHYL SULFOXIDE, ... | | 著者 | Lee, C.-J, Zhou, P. | | 登録日 | 2010-11-30 | | 公開日 | 2011-01-19 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Syntheses, structures and antibiotic activities of LpxC inhibitors based on the diacetylene scaffold.
Bioorg.Med.Chem., 19, 2011
|
|
