1WWL
 
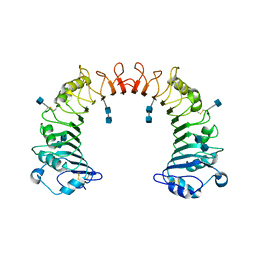 | | Crystal structure of CD14 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Monocyte differentiation antigen CD14 | | Authors: | Kim, J.-I, Lee, C.J, Jin, M.S, Lee, C.-H, Paik, S.-G, Lee, H, Lee, J.-O. | | Deposit date: | 2005-01-06 | | Release date: | 2005-02-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of CD14 and Its Implications for Lipopolysaccharide Signaling
J.Biol.Chem., 280, 2005
|
|
1EFN
 
 | |
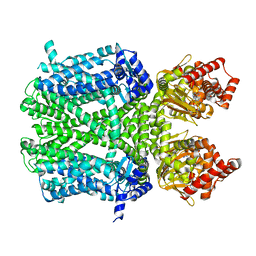
8WZX
 
 | | Cryo-EM structure of the hamster prion 23-144 fibril at pH 3.7 | | Descriptor: | Major prion protein | | Authors: | Lee, C.-H, Saw, J.-E, Chen, E, Wang, C.-H, Chen, R. | | Deposit date: | 2023-11-02 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | The Positively Charged Cluster in the N-terminal Disordered Region may Affect Prion Protein Misfolding: Cryo-EM Structure of Hamster PrP(23-144) Fibrils.
J.Mol.Biol., 436, 2024
|
|
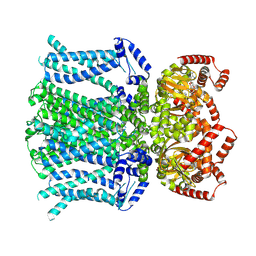
6UQF
 
 | | Human HCN1 channel in a hyperpolarized conformation | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, MERCURY (II) ION, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Lee, C.-H, MacKinnon, R. | | Deposit date: | 2019-10-19 | | Release date: | 2019-12-11 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Voltage Sensor Movements during Hyperpolarization in the HCN Channel.
Cell, 179, 2019
|
|
6UQG
 
 | | Human HCN1 channel Y289D mutant | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Lee, C.-H, MacKinnon, R. | | Deposit date: | 2019-10-19 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Voltage Sensor Movements during Hyperpolarization in the HCN Channel.
Cell, 179, 2019
|
|
5U6P
 
 | |
5U6O
 
 | |
1AYA
 
 | |
1AYB
 
 | |
1AQC
 
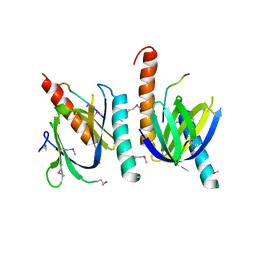 | | X11 PTB DOMAIN-10MER PEPTIDE COMPLEX | | Descriptor: | PEPTIDE, X11 | | Authors: | Lee, C.-H, Zhang, Z, Kuriyan, J. | | Deposit date: | 1997-07-28 | | Release date: | 1997-12-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Sequence-specific recognition of the internalization motif of the Alzheimer's amyloid precursor protein by the X11 PTB domain.
EMBO J., 16, 1997
|
|
1AYC
 
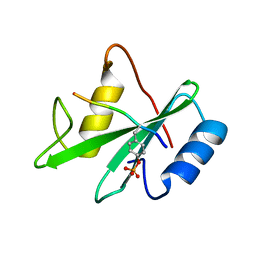 | |
1AYD
 
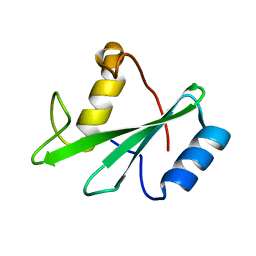 | |
1X11
 
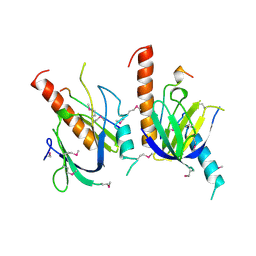 | | X11 PTB DOMAIN | | Descriptor: | 13-MER PEPTIDE, X11 | | Authors: | Lee, C.-H, Zhang, Z, Kuriyan, J. | | Deposit date: | 1997-07-28 | | Release date: | 1998-01-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sequence-specific recognition of the internalization motif of the Alzheimer's amyloid precursor protein by the X11 PTB domain.
EMBO J., 16, 1997
|
|
4TLL
 
 | | Crystal structure of GluN1/GluN2B NMDA receptor, structure 1 | | Descriptor: | 1-AMINOCYCLOPROPANECARBOXYLIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(1R,2S)-3-(4-benzylpiperidin-1-yl)-1-hydroxy-2-methylpropyl]phenol, ... | | Authors: | Gouaux, E, Lee, C.-H, Lu, W. | | Deposit date: | 2014-05-30 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | NMDA receptor structures reveal subunit arrangement and pore architecture.
Nature, 511, 2014
|
|
4TLM
 
 | | Crystal structure of GluN1/GluN2B NMDA receptor, structure 2 | | Descriptor: | 1-AMINOCYCLOPROPANECARBOXYLIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(1R,2S)-3-(4-benzylpiperidin-1-yl)-1-hydroxy-2-methylpropyl]phenol, ... | | Authors: | Gouaux, E, Lee, C.-H, Lu, W. | | Deposit date: | 2014-05-30 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.77 Å) | | Cite: | NMDA receptor structures reveal subunit arrangement and pore architecture.
Nature, 511, 2014
|
|
1JJG
 
 | | Solution Structure of Myxoma Virus Protein M156R | | Descriptor: | M156R | | Authors: | Ramelot, T.A, Cort, J.R, Yee, A.A, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-07-05 | | Release date: | 2002-03-06 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Myxoma virus immunomodulatory protein M156R is a structural mimic of eukaryotic translation initiation factor eIF2alpha.
J.Mol.Biol., 322, 2002
|
|
7YAT
 
 | | CryoEM tetra protofilament structure of the hamster prion 108-144 fibril | | Descriptor: | Major prion protein | | Authors: | Chen, E.H.-L, Kao, S.-W, Lee, C.-H, Huang, J.Y.C, Chen, R.P.-Y, Wu, K.-P. | | Deposit date: | 2022-06-28 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | 2.2 angstrom Cryo-EM Tetra-Protofilament Structure of the Hamster Prion 108-144 Fibril Reveals an Ordered Water Channel in the Center.
J.Am.Chem.Soc., 144, 2022
|
|
8ZMR
 
 | | Vesamicol-bound VAChT | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Vesicular acetylcholine transporter,DARPinoff7, vesamicol | | Authors: | Zhang, Z, Zhang, Y, Dai, F, Zhang, Y.X, Lee, C.-H. | | Deposit date: | 2024-05-23 | | Release date: | 2024-06-19 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into VAChT neurotransmitter recognition and inhibition.
Cell Res., 34, 2024
|
|
8ZMS
 
 | | Acetylcholine-bound VAChT | | Descriptor: | ACETYLCHOLINE, Maltose/maltodextrin-binding periplasmic protein,Vesicular acetylcholine transporter,DARPinoff7 | | Authors: | Zhang, Z, Zhang, Y, Dai, F, Zhang, Y.X, Lee, C.-H. | | Deposit date: | 2024-05-23 | | Release date: | 2024-06-19 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into VAChT neurotransmitter recognition and inhibition.
Cell Res., 34, 2024
|
|
