9OBP
 
 | | Cryo-EM structure of alpha-synuclein filaments derived from the frontal cortex of the case of atypical multiple system atrophy | | Descriptor: | Alpha-synuclein, Unidentified protein | | Authors: | Enomoto, M, Martinez-Valbuena, I, Forrest, S.L, Xu, X, Munhoz, R, Li, J, Rogaeva, E, Lang, A.E, Kovacs, G.G. | | Deposit date: | 2025-04-23 | | Release date: | 2025-06-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Lewy-MSA hybrid fold drives distinct neuronal alpha-synuclein pathology.
Commun Biol, 8, 2025
|
|
4O9X
 
 | | Crystal Structure of TcdB2-TccC3 | | Descriptor: | MERCURY (II) ION, TcdB2, TccC3 | | Authors: | Meusch, D, Gatsogiannis, C, Efremov, R.G, Lang, A.E, Hofnagel, O, Vetter, I.R, Aktories, K, Raunser, S. | | Deposit date: | 2014-01-03 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Mechanism of Tc toxin action revealed in molecular detail.
Nature, 508, 2014
|
|
4O9Y
 
 | | Crystal Structure of TcdA1 | | Descriptor: | TcdA1 | | Authors: | Meusch, D, Gatsogiannis, C, Efremov, R.G, Lang, A.E, Hofnagel, O, Vetter, I.R, Aktories, K, Raunser, S. | | Deposit date: | 2014-01-03 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.502 Å) | | Cite: | Mechanism of Tc toxin action revealed in molecular detail.
Nature, 508, 2014
|
|
9E9X
 
 | | Cryo-EM structure of alpha-synuclein filaments derived from the temporal cortex of the case of atypical multiple system atrophy | | Descriptor: | Alpha-synuclein, unknown protein deposit 1, unknown protein deposit 2 | | Authors: | Enomoto, M, Martinez-Valbuena, I, Forrest, S.L, Xu, X, Munhoz, R, Li, J, Rogaeva, E, Lang, A.E, Kovacs, G.G. | | Deposit date: | 2024-11-09 | | Release date: | 2025-06-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Lewy-MSA hybrid fold drives distinct neuronal alpha-synuclein pathology.
Commun Biol, 8, 2025
|
|
7P6M
 
 | | Hydrogenated refolded hen egg-white lysozyme | | Descriptor: | ACETATE ION, Lysozyme C, NITRATE ION | | Authors: | Ramos, J, Laux, V, Haertlein, M, Forsyth, V.T, Mossou, E, Larsen, S, Langkilde, A.E. | | Deposit date: | 2021-07-16 | | Release date: | 2021-12-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | The impact of folding modes and deuteration on the atomic resolution structure of hen egg-white lysozyme.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
8RLG
 
 | | Perdeuterated hen egg-white lysozyme at room temperature | | Descriptor: | ACETATE ION, Lysozyme C, NITRATE ION | | Authors: | Ramos, J, Laux, V, Mason, S.A, Lemee, M, Bowler, M, Diederichs, K, Haertlein, M, Forsyth, V.T, Mossou, E, Larsen, S, Langkilde, A.E. | | Deposit date: | 2024-01-03 | | Release date: | 2024-11-06 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Structure and dynamics of the active site of hen egg-white lysozyme from atomic resolution neutron crystallography.
Structure, 33, 2025
|
|
7AVE
 
 | | Perdeuterated refolded hen egg-white lysozyme at 100 K | | Descriptor: | ACETATE ION, Lysozyme C, NITRATE ION | | Authors: | Ramos, J, Laux, V, Haertlein, M, Erba Boeri, E, Forsyth, V.T, Larsen, S, Mossou, E, Langkilde, A.E. | | Deposit date: | 2020-11-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Structural insights into protein folding, stability and activity using in vivo perdeuteration of hen egg-white lysozyme.
Iucrj, 8, 2021
|
|
7AVF
 
 | | Triclinic hydrogenated hen egg-white lysozyme at 100 K (control) | | Descriptor: | ACETATE ION, Lysozyme, NITRATE ION | | Authors: | Ramos, J, Laux, V, Haertlein, M, Erba Boeri, E, Forsyth, V.T, Mossou, E, Larsen, S, Langkilde, A.E. | | Deposit date: | 2020-11-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural insights into protein folding, stability and activity using in vivo perdeuteration of hen egg-white lysozyme.
Iucrj, 8, 2021
|
|
7AVG
 
 | | Perdeuterated hen egg-white lysozyme at 100 K | | Descriptor: | ACETATE ION, Lysozyme, NITRATE ION | | Authors: | Ramos, J, Laux, V, Haertlein, M, Erba Boeri, E, Forsyth, V.T, Mossou, E, Larsen, S, Langkilde, A.E. | | Deposit date: | 2020-11-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural insights into protein folding, stability and activity using in vivo perdeuteration of hen egg-white lysozyme.
Iucrj, 8, 2021
|
|
8RLF
 
 | | Hydrogenated hen egg-white lysozyme at room temperature | | Descriptor: | ACETATE ION, Lysozyme C, NITRATE ION | | Authors: | Ramos, J, Laux, V, Mason, S.A, Lemee, M, Bowler, M, Diederichs, K, Haertlein, M, Forsyth, V.T, Mossou, E, Larsen, S, Langkilde, A.E. | | Deposit date: | 2024-01-03 | | Release date: | 2024-11-06 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Structure and dynamics of the active site of hen egg-white lysozyme from atomic resolution neutron crystallography.
Structure, 33, 2025
|
|
8RLH
 
 | | Neutron structure of hydrogenated hen egg-white lysozyme at room temperature | | Descriptor: | ACETATE ION, Lysozyme C, NITRATE ION | | Authors: | Ramos, J, Laux, V, Mason, S.A, Lemee, M, Bowler, M, Diederichs, K, Haertlein, M, Forsyth, V.T, Mossou, E, Larsen, S, Langkilde, A.E. | | Deposit date: | 2024-01-03 | | Release date: | 2024-11-06 | | Last modified: | 2025-01-29 | | Method: | NEUTRON DIFFRACTION (1.1 Å) | | Cite: | Structure and dynamics of the active site of hen egg-white lysozyme from atomic resolution neutron crystallography.
Structure, 33, 2025
|
|
8RLI
 
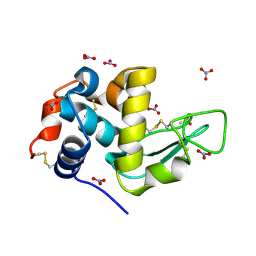 | | Neutron structure of perdeuterated hen egg-white lysozyme at room temperature | | Descriptor: | ACETATE ION, Lysozyme C, NITRATE ION | | Authors: | Ramos, J, Laux, V, Mason, S.A, Lemee, M, Bowler, M, Diederichs, K, Haertlein, M, Forsyth, V.T, Mossou, E, Larsen, S, Langkilde, A.E. | | Deposit date: | 2024-01-03 | | Release date: | 2024-11-06 | | Last modified: | 2025-01-29 | | Method: | NEUTRON DIFFRACTION (0.91 Å) | | Cite: | Structure and dynamics of the active site of hen egg-white lysozyme from atomic resolution neutron crystallography.
Structure, 33, 2025
|
|
6ZXL
 
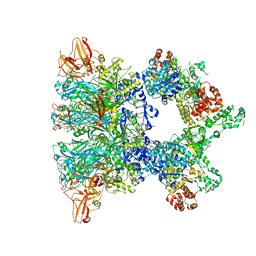 | | Fully-loaded anthrax lethal toxin in its heptameric pre-pore state and PA7LF(2+1A) arrangement | | Descriptor: | Lethal factor, Protective antigen | | Authors: | Quentin, D, Antoni, C, Gatsogiannis, C, Raunser, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of the fully-loaded asymmetric anthrax lethal toxin in its heptameric pre-pore state.
Plos Pathog., 16, 2020
|
|
6ZXK
 
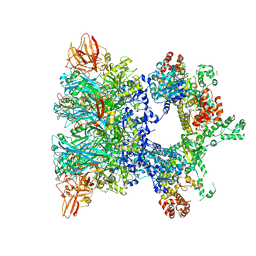 | | Fully-loaded anthrax lethal toxin in its heptameric pre-pore state and PA7LF(2+1B) arrangement | | Descriptor: | Lethal factor, Protective antigen | | Authors: | Quentin, D, Antoni, C, Gatsogiannis, C, Raunser, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the fully-loaded asymmetric anthrax lethal toxin in its heptameric pre-pore state.
Plos Pathog., 16, 2020
|
|
6ZXJ
 
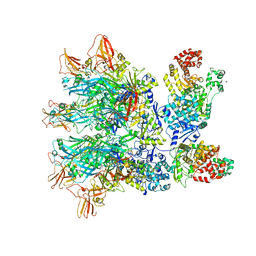 | | Fully-loaded anthrax lethal toxin in its heptameric pre-pore state, in which the third lethal factor is masked out (PA7LF3-masked) | | Descriptor: | Lethal factor, Protective antigen | | Authors: | Quentin, D, Antoni, C, Gatsogiannis, C, Raunser, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of the fully-loaded asymmetric anthrax lethal toxin in its heptameric pre-pore state.
Plos Pathog., 16, 2020
|
|
6RU2
 
 | | Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-methyl-glucuronoyl methylesterase, ... | | Authors: | Ernst, H.A, Mosbech, C, Langkilde, A, Westh, P, Meyer, A, Agger, J.W, Larsen, S. | | Deposit date: | 2019-05-27 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The structural basis of fungal glucuronoyl esterase activity on natural substrates.
Nat Commun, 11, 2020
|
|
6RV8
 
 | | Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor covalent complex with the aldouronic acid UXXR | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-Xylitol, ... | | Authors: | Ernst, H.A, Mosbech, C, Langkilde, A, Westh, P, Meyer, A, Agger, J.W, Larsen, S. | | Deposit date: | 2019-05-31 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structural basis of fungal glucuronoyl esterase activity on natural substrates.
Nat Commun, 11, 2020
|
|
6RTV
 
 | | Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-methyl-glucuronoyl methylesterase, ... | | Authors: | Ernst, H.A, Mosbech, C, Langkilde, A, Westh, P, Meyer, A, Agger, J.W, Larsen, S. | | Deposit date: | 2019-05-27 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The structural basis of fungal glucuronoyl esterase activity on natural substrates.
Nat Commun, 11, 2020
|
|
6RV9
 
 | | Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid XUXXR | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-[beta-D-xylopyranose-(1-4)]beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-Xylitol, ... | | Authors: | Ernst, H.A, Mosbech, C, Langkilde, A, Westh, P, Meyer, A, Agger, J.W, Larsen, S. | | Deposit date: | 2019-05-31 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The structural basis of fungal glucuronoyl esterase activity on natural substrates.
Nat Commun, 11, 2020
|
|
6RV7
 
 | | Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid UXXR | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-Xylitol, ... | | Authors: | Ernst, H.A, Mosbech, C, Langkilde, A, Westh, P, Meyer, A, Agger, J.W, Larsen, S. | | Deposit date: | 2019-05-31 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The structural basis of fungal glucuronoyl esterase activity on natural substrates.
Nat Commun, 11, 2020
|
|
6RU1
 
 | | Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid Um4X | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ... | | Authors: | Ernst, H.A, Mosbech, C, Langkilde, A, Westh, P, Meyer, A, Agger, J.W, Larsen, S. | | Deposit date: | 2019-05-27 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | The structural basis of fungal glucuronoyl esterase activity on natural substrates.
Nat Commun, 11, 2020
|
|
9EOY
 
 | | Structure of Thr354Asn, Glu355Gln, Thr412Asn, Ile414Met, Ile464His, and Phe467Met mutant human CaMKII alpha hub bound to PIPA | | Descriptor: | 2-[6-(4-chlorophenyl)imidazo[1,2-b]pyridazin-2-yl]ethanoic acid, ACETATE ION, Calcium/calmodulin-dependent protein kinase type II subunit alpha, ... | | Authors: | Narayanan, D, Larsen, A.S.G, Solbak, S.M.O, Wellendorph, P, Gee, C.L, Kastrup, J.S. | | Deposit date: | 2024-03-15 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ligand-induced CaMKII alpha hub Trp403 flip, hub domain stacking, and modulation of kinase activity.
Protein Sci., 33, 2024
|
|
