1WM8
 
 | | Solution Structure of BmP03 from the Venom of Scorpion Buthus martensii Karsch, 10 structures | | Descriptor: | Neurotoxin BmP03 | | Authors: | Wu, H, He, F, Li, Y, Wu, G, Cao, C, Chen, X. | | Deposit date: | 2004-07-05 | | Release date: | 2004-07-27 | | Last modified: | 2018-04-18 | | Method: | SOLUTION NMR | | Cite: | Three-Dimensional Structure of BmP03 from Venom of Scorpion Buthus martensii Karsch
Acta Chim.Sinica, 58, 2000
|
|
3DKM
 
 | | Crystal structure of the HECTD1 CPH domain | | Descriptor: | E3 ubiquitin-protein ligase HECTD1 | | Authors: | Walker, J.R, Qiu, L, Li, Y, Bountra, C, Wolkstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-06-25 | | Release date: | 2008-08-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the CPH domain of the E3 ubiquitin-protein ligase HECTD1.
To be Published
|
|
1WT8
 
 | | Solution Structure of BmP08 from the Venom of Scorpion Buthus martensii Karsch, 20 structures | | Descriptor: | Neurotoxin BmK X | | Authors: | Wu, H, Chen, X, Tong, X, Li, Y, Zhang, N, Wu, G. | | Deposit date: | 2004-11-17 | | Release date: | 2005-04-19 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmP08, a novel short-chain scorpion toxin from Buthus martensi Karsch.
Biochem.Biophys.Res.Commun., 330, 2005
|
|
1WM7
 
 | | Solution Structure of BmP01 from the Venom of Scorpion Buthus martensii Karsch, 9 structures | | Descriptor: | Neurotoxin BmP01 | | Authors: | Wu, G, Li, Y, Wei, D, He, F, Jiang, S, Hu, G, Wu, H, Chen, X. | | Deposit date: | 2004-07-05 | | Release date: | 2004-07-27 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of BmP01 from the Venom of Scorpion Buthus martensii Karsch
Biochem.Biophys.Res.Commun., 276, 2000
|
|
3E19
 
 | | Crystal Structure of Iron Uptake Regulatory Protein (FeoA) Solved by Sulfur SAD in a Monoclinic Space Group | | Descriptor: | FeoA, GLYCEROL, PHOSPHATE ION | | Authors: | Hughes, R.C, Li, Y, Wang, B.-C, Liu, Z.-J, Ng, J.D. | | Deposit date: | 2008-08-02 | | Release date: | 2008-12-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic Structure Determination of Iron Uptake Regulatory
Protein (FeoA) by Sulfur SAD in a Monoclinic Space Group
To be Published
|
|
1X9M
 
 | | T7 DNA polymerase in complex with an N-2-acetylaminofluorene-adducted DNA | | Descriptor: | 5'-D(*CP*CP*CP*(8FG)P*AP*TP*CP*AP*CP*AP*CP*TP*AP*CP*CP*AP*AP*TP*CP*AP*CP*TP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*AP*GP*TP*GP*AP*TP*TP*GP*GP*TP*AP*GP*TP*GP*TP*GP*AP*(2DT))-3', DNA polymerase, ... | | Authors: | Dutta, S, Li, Y, Johnson, D, Dzantiev, L, Richardson, C.C, Romano, L.J, Ellenberger, T. | | Deposit date: | 2004-08-23 | | Release date: | 2004-10-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of 2-acetylaminofluorene and 2-aminofluorene in complex with T7 DNA polymerase reveal mechanisms of mutagenesis.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
7T39
 
 | | Co-crystal structure of human PRMT9 in complex with MT221 inhibitor | | Descriptor: | 7-[5-S-(4-{[(2-ethylpyridin-3-yl)methyl]amino}butyl)-5-thio-beta-D-ribofuranosyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Protein arginine N-methyltransferase 9 | | Authors: | Zeng, H, Dong, A, Hutchinson, A, Seitova, A, Li, Y, Gao, Y.D, Schneider, S, Siliphaivanh, P, Sloman, D, Nicholson, B, Fischer, C, Hicks, J, Brown, P.J, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-12-07 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Co-crystal structure of human PRMT9 in complex with MT221 inhibitor
To Be Published
|
|
1X9W
 
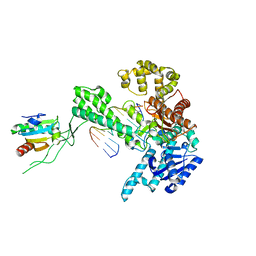 | | T7 DNA polymerase in complex with a primer/template DNA containing a disordered N-2 aminofluorene on the template, crystallized with dideoxy-ATP as the incoming nucleotide. | | Descriptor: | 5'-D(*CP*CP*CP*(AFG)*AP*TP*CP*AP*CP*AP*CP*TP*AP*CP*CP*AP*AP*TP*CP*AP*CP*TP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*AP*GP*TP*GP*AP*TP*T*GP*GP*T*AP*GP*TP*GP*TP*GP*AP*(2DT))-3', DNA polymerase, ... | | Authors: | Dutta, S, Li, Y, Johnson, D, Dzantiev, L, Richardson, C.C, Romano, L.J, Ellenberger, T. | | Deposit date: | 2004-08-24 | | Release date: | 2004-10-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of 2-acetylaminofluorene and 2-aminofluorene in complex with T7 DNA polymerase reveal mechanisms of mutagenesis.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1XC5
 
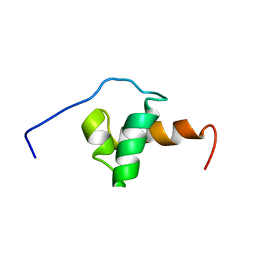 | | Solution Structure of the SMRT Deacetylase Activation Domain | | Descriptor: | Nuclear receptor corepressor 2 | | Authors: | Codina, A, Love, J.D, Li, Y, Lazar, M.A, Neuhaus, D, Schwabe, J.W.R. | | Deposit date: | 2004-09-01 | | Release date: | 2005-05-03 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the interaction and activation of histone deacetylase 3 by nuclear receptor corepressors
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1X9S
 
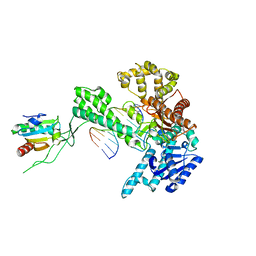 | | T7 DNA polymerase in complex with a primer/template DNA containing a disordered N-2 aminofluorene on the template, crystallized with dideoxy-CTP as the incoming nucleotide. | | Descriptor: | 5'-D(*CP*CP*CP*(AFG)P*AP*TP*CP*AP*CP*AP*CP*TP*AP*CP*CP*AP*AP*TP*CP*AP*CP*TP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*AP*GP*TP*GP*AP*TP*T*GP*GP*TP*AP*GP*TP*GP*TP*GP*AP*(2DT))-3', DNA polymerase, ... | | Authors: | Dutta, S, Li, Y, Johnson, D, Dzantiev, L, Richardson, C.C, Romano, L.J, Ellenberger, T. | | Deposit date: | 2004-08-24 | | Release date: | 2004-10-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of 2-acetylaminofluorene and 2-aminofluorene in complex with T7 DNA polymerase reveal mechanisms of mutagenesis.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1XLS
 
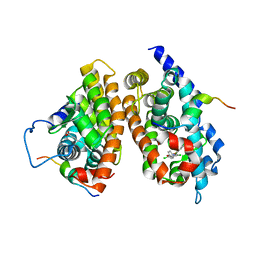 | | Crystal structure of the mouse CAR/RXR LBD heterodimer bound to TCPOBOP and 9cRA and a TIF2 peptide containg the third LXXLL motifs | | Descriptor: | (9cis)-retinoic acid, 3,5-DICHLORO-2-{4-[(3,5-DICHLOROPYRIDIN-2-YL)OXY]PHENOXY}PYRIDINE, Nuclear receptor coactivator 2, ... | | Authors: | Suino, K, peng, L, Reynolds, R, Li, Y, Cha, J.-Y, Repa, J.J, Kliewer, S.A, Xu, H.E. | | Deposit date: | 2004-09-30 | | Release date: | 2004-12-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | The nuclear xenobiotic receptor CAR: structural determinants of constitutive activation and heterodimerization.
Mol.Cell, 16, 2004
|
|
1XK7
 
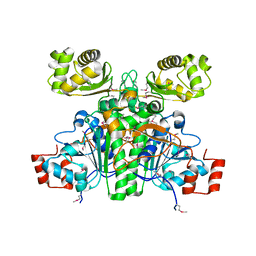 | | Crystal Structure- C2 form- of Escherichia coli Crotonobetainyl-CoA: carnitine CoA transferase (CaiB) | | Descriptor: | Crotonobetainyl-CoA:carnitine CoA-transferase | | Authors: | Rangarajan, E.S, Li, Y, Iannuzzi, P, Cygler, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2004-09-27 | | Release date: | 2005-03-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Escherichia coli Crotonobetainyl-CoA: Carnitine CoA-Transferase (CaiB) and Its Complexes with CoA and Carnitinyl-CoA.
Biochemistry, 44, 2005
|
|
4JUY
 
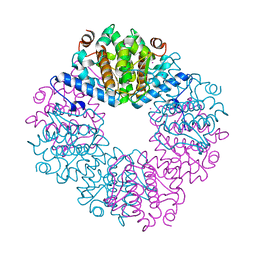 | | Crystal structure of the PUB domain of E3 ubiquitin ligase RNF31 | | Descriptor: | E3 ubiquitin-protein ligase RNF31, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Hu, J, Li, Y, Wernimont, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-25 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the PUB domain of E3 ubiquitin ligase RNF31
To be Published
|
|
1XVT
 
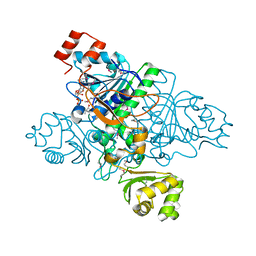 | | Crystal Structure of Native CaiB in complex with coenzyme A | | Descriptor: | COENZYME A, Crotonobetainyl-CoA:carnitine CoA-transferase | | Authors: | Rangarajan, E.S, Li, Y, Iannuzzi, P, Cygler, M, Matte, A. | | Deposit date: | 2004-10-28 | | Release date: | 2005-03-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Escherichia coli Crotonobetainyl-CoA: Carnitine CoA-Transferase (CaiB) and Its Complexes with CoA and Carnitinyl-CoA.
Biochemistry, 44, 2005
|
|
1XK6
 
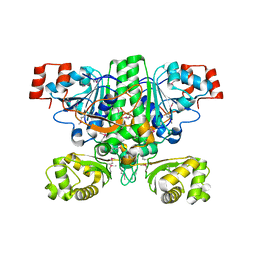 | | Crystal Structure- P1 form- of Escherichia coli Crotonobetainyl-CoA: carnitine CoA Transferase (CaiB) | | Descriptor: | Crotonobetainyl-CoA:carnitine CoA-transferase | | Authors: | Rangarajan, E.S, Li, Y, Iannuzzi, P, Cygler, M, Matte, A. | | Deposit date: | 2004-09-27 | | Release date: | 2005-03-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Escherichia coli Crotonobetainyl-CoA: Carnitine CoA-Transferase (CaiB) and Its Complexes with CoA and Carnitinyl-CoA.
Biochemistry, 44, 2005
|
|
1XVU
 
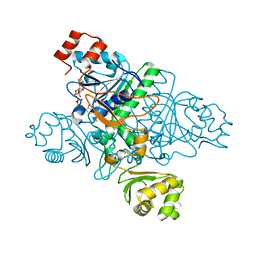 | | Crystal Structure of CaiB mutant D169A in complex with Coenzyme A | | Descriptor: | COENZYME A, Crotonobetainyl-CoA:carnitine CoA-transferase | | Authors: | Rangarajan, E.S, Li, Y, Iannuzzi, P, Cygler, M, Matte, A. | | Deposit date: | 2004-10-28 | | Release date: | 2005-03-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Escherichia coli Crotonobetainyl-CoA: Carnitine CoA-Transferase (CaiB) and Its Complexes with CoA and Carnitinyl-CoA.
Biochemistry, 44, 2005
|
|
1XVV
 
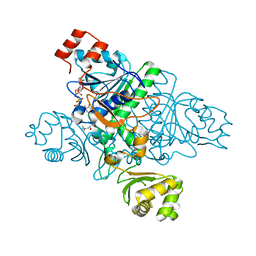 | | Crystal Structure of CaiB mutant D169A in complex with carnitinyl-CoA | | Descriptor: | Crotonobetainyl-CoA:carnitine CoA-transferase, L-CARNITINYL-COA INNER SALT | | Authors: | Rangarajan, E.S, Li, Y, Iannuzzi, P, Cygler, M, Matte, A. | | Deposit date: | 2004-10-28 | | Release date: | 2005-03-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Escherichia coli Crotonobetainyl-CoA: Carnitine CoA-Transferase (CaiB) and Its Complexes with CoA and Carnitinyl-CoA.
Biochemistry, 44, 2005
|
|
1Y00
 
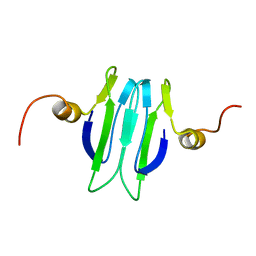 | | Solution structure of the Carbon Storage Regulator protein CsrA | | Descriptor: | Carbon storage regulator | | Authors: | Gutierrez, P, Li, Y, Osborne, M.J, Liu, Q, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2004-11-13 | | Release date: | 2005-06-21 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the carbon storage regulator protein CsrA from Escherichia coli.
J.Bacteriol., 187, 2005
|
|
3F0N
 
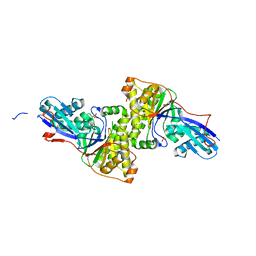 | | Mus Musculus Mevalonate Pyrophosphate Decarboxylase | | Descriptor: | MEVALONATE PYROPHOSPHATE DECARBOXYLASE, PHOSPHATE ION | | Authors: | Walker, J.R, Davis, T, Vesterberg, A, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-10-25 | | Release date: | 2008-11-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Mus Musculus Mevalonate Pyrophosphate Decarboxylase
To be Published
|
|
1Y6H
 
 | | Crystal structure of LIPDF | | Descriptor: | FORMIC ACID, GLYCINE, Peptide deformylase, ... | | Authors: | Zhou, Z, Song, X, Li, Y, Gong, W. | | Deposit date: | 2004-12-06 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unique structural characteristics of peptide deformylase from pathogenic bacterium Leptospira interrogans
J.Mol.Biol., 339, 2004
|
|
3FCX
 
 | | Crystal structure of human esterase D | | Descriptor: | CALCIUM ION, MAGNESIUM ION, S-formylglutathione hydrolase | | Authors: | Wu, D, Li, Y, Song, G, Zhang, D, Shaw, N, Liu, Z.J. | | Deposit date: | 2008-11-24 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human esterase D: a potential genetic marker of retinoblastoma
Faseb J., 23, 2009
|
|
3FL2
 
 | | Crystal structure of the ring domain of the E3 ubiquitin-protein ligase UHRF1 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-18 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the Ring Domain of the E3 Ubiquitin-Protein Ligase Uhrf1
To be Published
|
|
6IZT
 
 | | Crystal structure of Haemophilus Influenzae BamA POTRA3-5 | | Descriptor: | Outer membrane protein assembly factor BamA | | Authors: | Ma, X, Wang, Q, Li, Y, Tan, P, Wu, H, Wang, P, Dong, X, Hong, L, Meng, G. | | Deposit date: | 2018-12-20 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | How BamA recruits OMP substratesviapoly-POTRAs domain.
Faseb J., 33, 2019
|
|
6IZS
 
 | | Crystal structure of Haemophilus influenzae BamA POTRA4 | | Descriptor: | Outer membrane protein assembly factor BamA | | Authors: | Ma, X, Wang, Q, Li, Y, Tan, P, Wu, H, Wang, P, Dong, X, Hong, L, Meng, G. | | Deposit date: | 2018-12-20 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | How BamA recruits OMP substratesviapoly-POTRAs domain.
Faseb J., 33, 2019
|
|
1YNI
 
 | | Crystal Structure of N-Succinylarginine Dihydrolase, AstB, bound to Substrate and Product, an Enzyme from the Arginine Catabolic Pathway of Escherichia coli | | Descriptor: | N~2~-(3-CARBOXYPROPANOYL)-L-ARGININE, POTASSIUM ION, Succinylarginine Dihydrolase | | Authors: | Tocilj, A, Schrag, J.D, Li, Y, Schneider, B.L, Reitzer, L, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2005-01-24 | | Release date: | 2005-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of N-succinylarginine dihydrolase AstB, bound to substrate and product, an enzyme from the arginine catabolic pathway of Escherichia coli.
J.Biol.Chem., 280, 2005
|
|
