7U3G
 
 | | GID4 in complex with compound 67 | | Descriptor: | (1R)-1-phenyl-1,2,3,4-tetrahydroisoquinolin-5-amine, Glucose-induced degradation protein 4 homolog | | Authors: | Chana, C.K, Sicheri, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Discovery and Structural Characterization of Small Molecule Binders of the Human CTLH E3 Ligase Subunit GID4.
J.Med.Chem., 65, 2022
|
|
7U3K
 
 | | GID4 in complex with compound 89 | | Descriptor: | GLYCEROL, Glucose-induced degradation protein 4 homolog, N-butylglycyl-4-tert-butyl-D-phenylalanyl-3-methoxy-N-methyl-L-phenylalaninamide | | Authors: | Chana, C.K, Sicheri, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Discovery and Structural Characterization of Small Molecule Binders of the Human CTLH E3 Ligase Subunit GID4.
J.Med.Chem., 65, 2022
|
|
7UKZ
 
 | |
7U3I
 
 | | GID4 in complex with compound 16 | | Descriptor: | 3-{[(5S)-5-methyl-4,5-dihydro-1,3-thiazol-2-yl]amino}phenol, GLYCEROL, Glucose-induced degradation protein 4 homolog | | Authors: | Chana, C.K, Sicheri, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | Discovery and Structural Characterization of Small Molecule Binders of the Human CTLH E3 Ligase Subunit GID4.
J.Med.Chem., 65, 2022
|
|
7U3J
 
 | | GID4 in complex with compound 88 | | Descriptor: | (2S)-2-{[(2S)-2-({N-[(2,4-dimethoxyphenyl)methyl]glycyl}amino)-2-(thiophen-2-yl)acetyl]amino}-N-methyl-4-phenylbutanamide, GLYCEROL, Glucose-induced degradation protein 4 homolog | | Authors: | Chana, C.K, Sicheri, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.642 Å) | | Cite: | Discovery and Structural Characterization of Small Molecule Binders of the Human CTLH E3 Ligase Subunit GID4.
J.Med.Chem., 65, 2022
|
|
7L84
 
 | |
6D4P
 
 | | Ube2D1 in complex with ubiquitin variant Ubv.D1.1 | | Descriptor: | Ubiquitin Variant Ubv.D1.1, Ubiquitin-conjugating enzyme E2 D1 | | Authors: | Ceccarelli, D.F, Garg, P, Sidhu, S, Sicheri, F. | | Deposit date: | 2018-04-18 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural and Functional Analysis of Ubiquitin-based Inhibitors That Target the Backsides of E2 Enzymes.
J.Mol.Biol., 432, 2020
|
|
6D68
 
 | | Ube2G1 in complex with ubiquitin variant Ubv.G1.1 | | Descriptor: | Ubiquitin-conjugating enzyme E2 G1, Ubv.G1.1 | | Authors: | Ceccarelli, D.F, Garg, P, Sidhu, S, Sicheri, F. | | Deposit date: | 2018-04-20 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural and Functional Analysis of Ubiquitin-based Inhibitors That Target the Backsides of E2 Enzymes.
J.Mol.Biol., 432, 2020
|
|
6D6I
 
 | | Ube2V1 in complex with ubiquitin variant Ubv.V1.1 and Ube2N/Ubc13 | | Descriptor: | Ubiquitin-conjugating enzyme E2 N, Ubiquitin-conjugating enzyme E2 variant 1, Ubv.V1.1 | | Authors: | Ceccarelli, D.F, Garg, P, Keszei, A, Sidhu, S, Sicheri, F. | | Deposit date: | 2018-04-21 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Structural and Functional Analysis of Ubiquitin-based Inhibitors That Target the Backsides of E2 Enzymes.
J.Mol.Biol., 432, 2020
|
|
6C16
 
 | |
6BYH
 
 | |
6BXI
 
 | | X-ray crystal structure of NDR1 kinase domain | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase 38 | | Authors: | Xiong, S, Sicheri, F. | | Deposit date: | 2017-12-18 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Auto-Inhibition of the NDR1 Kinase Domain by an Atypically Long Activation Segment.
Structure, 26, 2018
|
|
6BVA
 
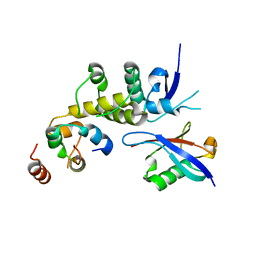 | |
6DRM
 
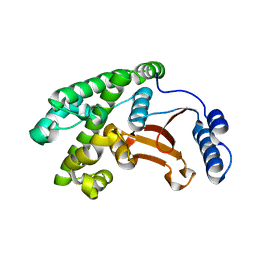 | | OTU domain of Fam105A | | Descriptor: | Inactive ubiquitin thioesterase FAM105A | | Authors: | Ceccarelli, D.F, Sicheri, F, Cordes, S. | | Deposit date: | 2018-06-12 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | FAM105A/OTULINL Is a Pseudodeubiquitinase of the OTU-Class that Localizes to the ER Membrane.
Structure, 27, 2019
|
|
