9ICJ
 
 | |
9ICE
 
 | |
9ICI
 
 | |
9ICB
 
 | |
5EVO
 
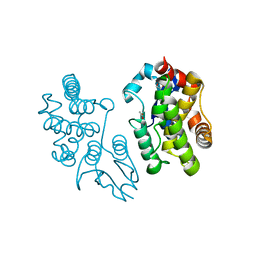 | | Structure of Dehydroascrobate Reductase from Pennisetum Americanum in complex with two non-native ligands, Acetate in the G-site and Glycerol in the H-site | | Descriptor: | ACETATE ION, Dehydroascorbate reductase, GLYCEROL | | Authors: | Kumar, A.O, Das, B.K, Arockiasamy, A. | | Deposit date: | 2015-11-20 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Non-native ligands define the active site of Pennisetum glaucum (L.) R. Br dehydroascorbate reductase.
Biochem.Biophys.Res.Commun., 473, 2016
|
|
5MU1
 
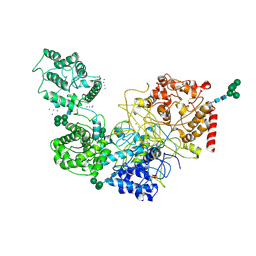 | | UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum soaked with K2PtI6 | | Descriptor: | CALCIUM ION, IODIDE ION, PLATINUM (II) ION, ... | | Authors: | Roversi, P, Caputo, A.T, Hill, J, Alonzi, D.S, Zitzmann, N. | | Deposit date: | 2017-01-11 | | Release date: | 2017-07-26 | | Last modified: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Interdomain conformational flexibility underpins the activity of UGGT, the eukaryotic glycoprotein secretion checkpoint.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5MZO
 
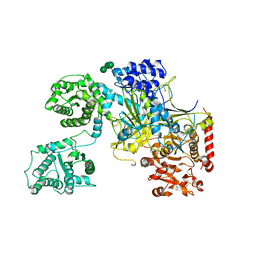 | | UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum (open conformation) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roversi, P, Caputo, A.T, Hill, J, Alonzi, D.S, Zitzmann, N. | | Deposit date: | 2017-02-01 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Interdomain conformational flexibility underpins the activity of UGGT, the eukaryotic glycoprotein secretion checkpoint.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5N2J
 
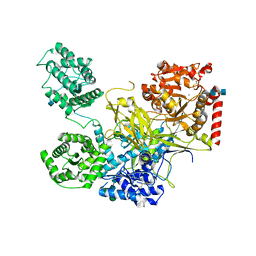 | | UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum (closed form) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roversi, P, Caputo, A.T, Hill, J, Alonzi, D.S, Zitzmann, N. | | Deposit date: | 2017-02-07 | | Release date: | 2017-07-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Interdomain conformational flexibility underpins the activity of UGGT, the eukaryotic glycoprotein secretion checkpoint.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5NV4
 
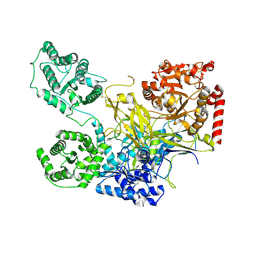 | | UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum double mutant D611C:G1050C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FORMIC ACID, UDP-glucose-glycoprotein glucosyltransferase-like protein, ... | | Authors: | Roversi, P, Caputo, A.T, Hill, J, Alonzi, D.S, Zitzmann, N. | | Deposit date: | 2017-05-03 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Interdomain conformational flexibility underpins the activity of UGGT, the eukaryotic glycoprotein secretion checkpoint.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8IVX
 
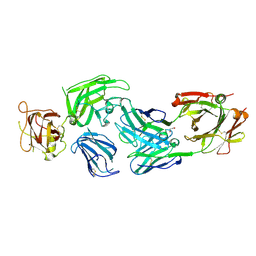 | | Crystal structure of NRP2 in complex with aNRP2-14 Fab fragment | | Descriptor: | 1,2-ETHANEDIOL, Heavy chain of antibody 14V4 Fab fragment, Light chain of antibody 14V4 Fab fragment, ... | | Authors: | Geng, Y, Zhai, L. | | Deposit date: | 2023-03-29 | | Release date: | 2023-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibition of VEGF binding to neuropilin-2 enhances chemosensitivity and inhibits metastasis in triple-negative breast cancer.
Sci Transl Med, 15, 2023
|
|
8IVW
 
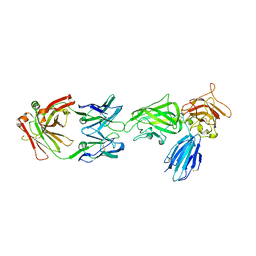 | | Crystal structure of NRP2 in complex with aNRP2-10 Fab fragment | | Descriptor: | Heavy chian of antibody 10V8 Fab fragment, Light chain of antibody 10V8 Fab fragment, Neuropilin-2 | | Authors: | Geng, Y, Zhai, L. | | Deposit date: | 2023-03-29 | | Release date: | 2023-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.205 Å) | | Cite: | Inhibition of VEGF binding to neuropilin-2 enhances chemosensitivity and inhibits metastasis in triple-negative breast cancer.
Sci Transl Med, 15, 2023
|
|
7BWE
 
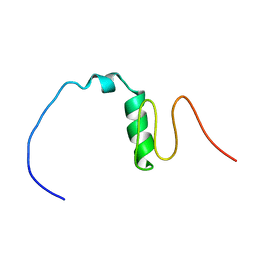 | | Consensus Chitin binding domain | | Descriptor: | Chitin binding beak protein 3 | | Authors: | Mohanram, H, Miserez, A. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a consensus chitin-binding domain revealed by solution NMR.
J.Struct.Biol., 213, 2021
|
|
7BWO
 
 | | Consensus chitin binding protein | | Descriptor: | Chitin binding beak protein 3 | | Authors: | Mohanram, H, Miserez, A. | | Deposit date: | 2020-04-15 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a consensus chitin-binding domain revealed by solution NMR.
J.Struct.Biol., 213, 2021
|
|
7DK1
 
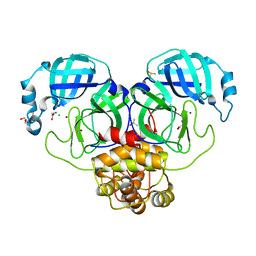 | | Crystal structure of Zinc bound SARS-CoV-2 main protease | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Sonkar, K.S, Panchariya, L, Kuila, S, Khan, W.A, Arockiasamy, A. | | Deposit date: | 2020-11-22 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Zinc 2+ ion inhibits SARS-CoV-2 main protease and viral replication in vitro.
Chem.Commun.(Camb.), 57, 2021
|
|
5F9O
 
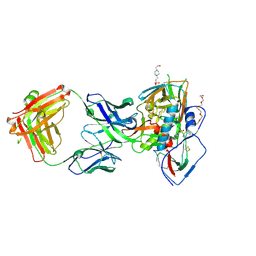 | | Crystal structure of broadly neutralizing VH1-46 germline-derived CD4-binding site-directed antibody CH235.09 in complex with HIV-1 clade A/E 93TH057 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CH235.09 Light chain, ... | | Authors: | Joyce, M.G, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Maturation Pathway from Germline to Broad HIV-1 Neutralizer of a CD4-Mimic Antibody.
Cell, 165, 2016
|
|
5F9W
 
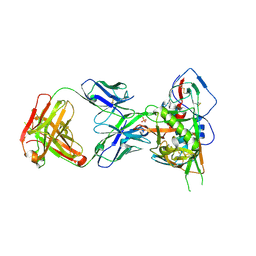 | | Crystal structure of broadly neutralizing VH1-46 germline-derived CD4-binding site-directed antibody CH235 in complex with HIV-1 clade A/E 93TH057 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of CH235-lineage antibody CH235, Light chain of CH235-lineage antibody CH235, ... | | Authors: | Chen, L, Zhou, T, Kwong, P.D. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8911 Å) | | Cite: | Maturation Pathway from Germline to Broad HIV-1 Neutralizer of a CD4-Mimic Antibody.
Cell, 165, 2016
|
|
5F96
 
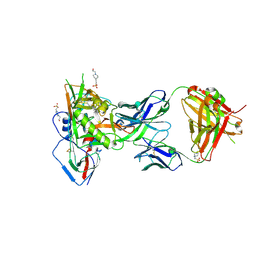 | |
2MI1
 
 | |
6HTN
 
 | | Structure of a fucose lectin from Kordia zhangzhouensis in complex with methyl-fucoside | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, ... | | Authors: | Varrot, A. | | Deposit date: | 2018-10-04 | | Release date: | 2019-01-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Architecture and Evolution of Blade Assembly in beta-propeller Lectins.
Structure, 27, 2019
|
|
6TS8
 
 | | Chaetomium thermophilum UDP-Glucose Glucosyl Transferase (UGGT) double cysteine mutant G177C/A786C. | | Descriptor: | UDP-glucose-glycoprotein glucosyltransferase-like protein | | Authors: | Roversi, P, Zitzmann, N, Ibba, R, Hensen, M, Chandran, A. | | Deposit date: | 2019-12-20 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | Clamping, bending, and twisting inter-domain motions in the misfold-recognizing portion of UDP-glucose: Glycoprotein glucosyltransferase.
Structure, 29, 2021
|
|
6TRT
 
 | | Chaetomium thermophilum UDP-Glucose Glucosyl Transferase (UGGT) double cysteine mutant S180C/T742C. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, TERBIUM(III) ION, UDP-glucose-glycoprotein glucosyltransferase-like protein, ... | | Authors: | Roversi, P, Zitzmann, N, Ibba, R, Hensen, M. | | Deposit date: | 2019-12-19 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.58 Å) | | Cite: | Clamping, bending, and twisting inter-domain motions in the misfold-recognizing portion of UDP-glucose: Glycoprotein glucosyltransferase.
Structure, 29, 2021
|
|
6TRF
 
 | | Chaetomium thermophilum UDP-Glucose Glucosyl Transferase (UGGT) purified from cells treated with kifunensine. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, UDP-glucose-glycoprotein glucosyltransferase-like protein, ... | | Authors: | Roversi, P, Zitzmann, N. | | Deposit date: | 2019-12-18 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.106 Å) | | Cite: | Clamping, bending, and twisting inter-domain motions in the misfold-recognizing portion of UDP-glucose: Glycoprotein glucosyltransferase.
Structure, 29, 2021
|
|
6TS2
 
 | | Truncated version of Chaetomium thermophilum UDP-Glucose Glucosyl Transferase (UGGT) lacking domain TRXL2 (417-650). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, UDP-glucose-glycoprotein glucosyltransferase-like protein,UDP-glucose-glycoprotein glucosyltransferase-like protein, ... | | Authors: | Roversi, P, Zitzmann, N. | | Deposit date: | 2019-12-19 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (5.74 Å) | | Cite: | Clamping, bending, and twisting inter-domain motions in the misfold-recognizing portion of UDP-glucose: Glycoprotein glucosyltransferase.
Structure, 29, 2021
|
|
3K5J
 
 | |
3KK7
 
 | |
