2F33
 
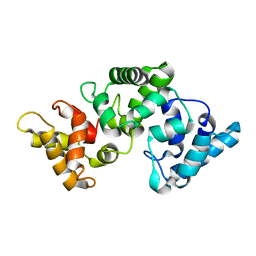 | | NMR solution structure of Ca2+-loaded calbindin D28K | | Descriptor: | Calbindin | | Authors: | Kojetin, D.J, Venters, R.A, Kordys, D.R, Thompson, R.J, Kumar, R, Cavanagh, J. | | Deposit date: | 2005-11-18 | | Release date: | 2006-07-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure, binding interface and hydrophobic transitions of Ca(2+)-loaded calbindin-D(28K).
Nat.Struct.Mol.Biol., 13, 2006
|
|
2G9B
 
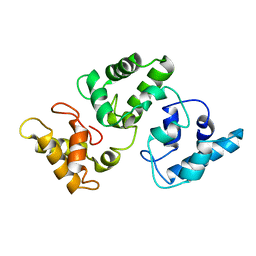 | | NMR solution structure of CA2+-loaded calbindin D28K | | Descriptor: | Calbindin | | Authors: | Kojetin, D.J, Venters, R.A, Kordys, D.R, Thompson, R.J, Kumar, R, Cavanagh, J. | | Deposit date: | 2006-03-06 | | Release date: | 2006-07-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure, binding interface and hydrophobic transitions of Ca(2+)-loaded calbindin-D(28K).
Nat.Struct.Mol.Biol., 13, 2006
|
|
2K77
 
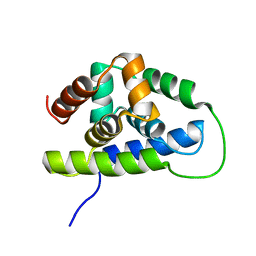 | | NMR solution structure of the Bacillus subtilis ClpC N-domain | | Descriptor: | Negative regulator of genetic competence clpC/mecB | | Authors: | Kojetin, D.J, McLaughlin, P.D, Thompson, R.J, Rance, M, Cavanagh, J. | | Deposit date: | 2008-08-04 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural and motional contributions of the Bacillus subtilis ClpC N-domain to adaptor protein interactions.
J.Mol.Biol., 387, 2009
|
|
4ZO1
 
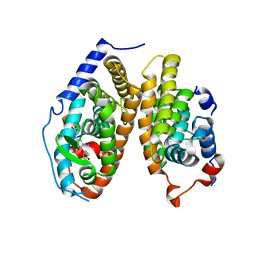 | | Crystal Structure of the T3-bound TR-beta Ligand-binding Domain in complex with RXR-alpha | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha, ... | | Authors: | Bruning, J.B, Kojetin, D.J, Matta-Camacho, E, Hughes, T.S, Srinivasan, S, Nwachukwu, J.C, Cavett, V, Nowak, J, Chalmers, M.J, Marciano, D.P, Kamenecka, T.M, Rance, M, Shulman, A.I, Mangelsdorf, D.J, Griffin, P.R, Nettles, K.W. | | Deposit date: | 2015-05-05 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.221 Å) | | Cite: | Structural mechanism for signal transduction in RXR nuclear receptor heterodimers.
Nat Commun, 6, 2015
|
|
8ZFQ
 
 | |
8ZFO
 
 | |
8ZFT
 
 | |
8ZFR
 
 | |
8ZFP
 
 | |
8ZFS
 
 | |
8ZFN
 
 | |
6ONI
 
 | |
2L7M
 
 | |
2L7F
 
 | |
6MD4
 
 | |
6MD1
 
 | |
6MCZ
 
 | |
2RO4
 
 | | RDC-refined Solution Structure of the N-terminal DNA Recognition Domain of the Bacillus subtilis Transition-state Regulator AbrB | | Descriptor: | Transition state regulatory protein abrB | | Authors: | Sullivan, D.M, Bobay, B.G, Kojetin, D.J, Thompson, R.J, Rance, M, Strauch, M.A, Cavanagh, J. | | Deposit date: | 2008-03-08 | | Release date: | 2008-11-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Insights into the nature of DNA binding of AbrB-like transcription factors
Structure, 16, 2008
|
|
2RO5
 
 | | RDC-refined solution structure of the N-terminal DNA recognition domain of the Bacillus subtilis transition-state regulator SpoVT | | Descriptor: | Stage V sporulation protein T | | Authors: | Sullivan, D.M, Bobay, B.G, Kojetin, D.J, Thompson, R.J, Rance, M, Strauch, M.A, Cavanagh, J. | | Deposit date: | 2008-03-08 | | Release date: | 2008-11-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Insights into the nature of DNA binding of AbrB-like transcription factors
Structure, 16, 2008
|
|
5UGM
 
 | | Crystal Structure of Human PPARgamma Ligand Binding Domain in Complex with Edaglitazone | | Descriptor: | (5R)-5-({4-[2-(5-methyl-2-phenyl-1,3-oxazol-4-yl)ethoxy]-1-benzothiophen-7-yl}methyl)-1,3-thiazolidine-2,4-dione, Peroxisome proliferator-activated receptor gamma, nonanoic acid | | Authors: | Shang, J, Kojetin, D.J. | | Deposit date: | 2017-01-09 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cooperative cobinding of synthetic and natural ligands to the nuclear receptor PPAR gamma.
Elife, 7, 2018
|
|
4IWC
 
 | | Crystal Structure of the Estrogen Receptor alpha Ligand-binding Domain in Complex with a Dynamic Thiophene-derivative | | Descriptor: | 4,4'-thiene-2,5-diylbis(3-methylphenol), Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Parent, A.A, Cavett, V, Nowak, J, Hughes, T.S, Kojetin, D.J, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2013-01-23 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Ligand binding dynamics rewire cellular signaling via Estrogen Receptor-alpha
Nat.Chem.Biol., 9, 2013
|
|
4IW6
 
 | | Crystal Structure of the Estrogen Receptor alpha Ligand-binding Domain in Complex with Constrained WAY-derivative, 7b | | Descriptor: | 4-[2-(but-3-en-1-yl)-7-(trifluoromethyl)-2H-indazol-3-yl]benzene-1,3-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Parent, A.A, Cavett, V, Nowak, J, Hughes, T.S, Kojetin, D.J, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2013-01-23 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Ligand binding dynamics rewire cellular signaling via Estrogen Receptor-alpha
Nat.Chem.Biol., 9, 2013
|
|
4IU7
 
 | | Crystal Structure of the Estrogen Receptor alpha Ligand-binding Domain in Complex with Constrained WAY-derivative, 2b | | Descriptor: | 4-[2-ethyl-7-(trifluoromethyl)-2H-indazol-3-yl]benzene-1,3-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Parent, A.A, Cavett, V, Nowak, J, Hughes, T.S, Kojetin, D.J, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2013-01-20 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Ligand binding dynamics rewire cellular signaling via Estrogen Receptor-alpha
Nat.Chem.Biol., 9, 2013
|
|
4IV4
 
 | | Crystal Structure of the Estrogen Receptor alpha Ligand-binding Domain in Complex with Constrained WAY-derivative, 5b | | Descriptor: | 4-[2-(2-methylpropyl)-7-(trifluoromethyl)-2H-indazol-3-yl]benzene-1,3-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Parent, A.A, Cavett, V, Nowak, J, Hughes, T.S, Kojetin, D.J, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2013-01-22 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ligand binding dynamics rewire cellular signaling via Estrogen Receptor-alpha
Nat.Chem.Biol., 9, 2013
|
|
4IV2
 
 | | Crystal Structure of the Estrogen Receptor alpha Ligand-binding Domain in Complex with Dynamic WAY-derivative, 5a | | Descriptor: | 4-[1-(2-methylpropyl)-7-(trifluoromethyl)-1H-indazol-3-yl]benzene-1,3-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Parent, A.A, Cavett, V, Nowak, J, Hughes, T.S, Kojetin, D.J, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2013-01-22 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Ligand binding dynamics rewire cellular signaling via Estrogen Receptor-alpha
Nat.Chem.Biol., 9, 2013
|
|
