3BEV
 
 | |
2A58
 
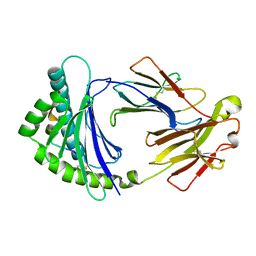
 | | Structure of 6,7-Dimethyl-8-ribityllumazine synthase from Schizosaccharomyces pombe mutant W27Y with bound riboflavin | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION, RIBOFLAVIN | | Authors: | Koch, M, Breithaupt, C, Gerhardt, S, Haase, I, Weber, S, Cushman, M, Huber, R, Bacher, A, Fischer, M. | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of charge transfer complex formation by riboflavin bound to 6,7-dimethyl-8-ribityllumazine synthase
Eur.J.Biochem., 271, 2004
|
|
2A57
 
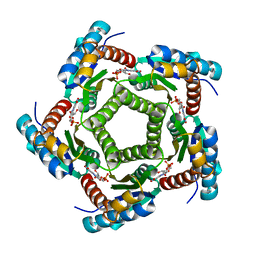 | | Structure of 6,7-Dimthyl-8-ribityllumazine synthase from Schizosaccharomyces pombe mutant W27Y with bound ligand 6-carboxyethyl-7-oxo-8-ribityllumazine | | Descriptor: | 3-[8-((2S,3S,4R)-2,3,4,5-TETRAHYDROXYPENTYL)-2,4,7-TRIOXO-1,3,8-TRIHYDROPTERIDIN-6-YL]PROPANOIC ACID, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Koch, M, Breithaupt, C, Gerhardt, S, Haase, I, Weber, S, Cushman, M, Huber, R, Bacher, A, Fischer, M. | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis of charge transfer complex formation by riboflavin bound to 6,7-dimethyl-8-ribityllumazine synthase
Eur.J.Biochem., 271, 2004
|
|
2A59
 
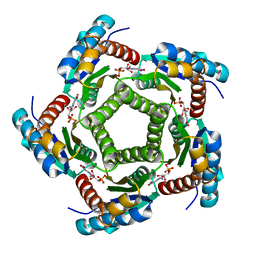 | | Structure of 6,7-Dimethyl-8-ribityllumazine synthase from Schizosaccharomyces pombe mutant W27Y with bound ligand 5-nitroso-6-ribitylamino-2,4(1H,3H)-pyrimidinedione | | Descriptor: | 5-NITROSO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Koch, M, Breithaupt, C, Gerhardt, S, Haase, I, Weber, S, Cushman, M, Huber, R, Bacher, A, Fischer, M. | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of charge transfer complex formation by riboflavin bound to 6,7-dimethyl-8-ribityllumazine synthase
Eur.J.Biochem., 271, 2004
|
|
1X9U
 
 | | Umecyanin from Horse Raddish- Crystal Structure of the reduced form | | Descriptor: | COPPER (II) ION, Umecyanin | | Authors: | Koch, M, Velarde, M, Harrison, M.D, Echt, S, Fischer, M, Messerschmidt, A, Dennison, C. | | Deposit date: | 2004-08-24 | | Release date: | 2005-03-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Oxidized and Reduced Stellacyanin from Horseradish Roots
J.Am.Chem.Soc., 127, 2005
|
|
1X9R
 
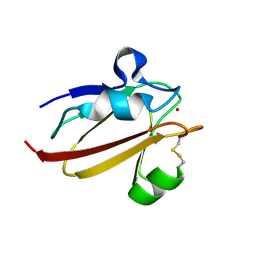 | | Umecyanin from Horse Raddish- Crystal Structure of the oxidised form | | Descriptor: | COPPER (II) ION, Umecyanin | | Authors: | Koch, M, Velarde, M, Harrison, M.D, Echt, S, Fischer, M, Messerschmidt, A, Dennison, C. | | Deposit date: | 2004-08-24 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Oxidized and Reduced Stellacyanin from Horseradish Roots
J.Am.Chem.Soc., 127, 2005
|
|
1SEZ
 
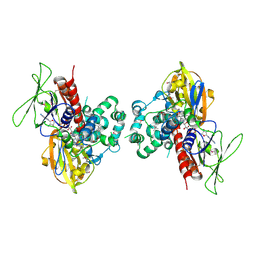 | | Crystal Structure of Protoporphyrinogen IX Oxidase | | Descriptor: | 2-{2-[4-(1,1,3,3-TETRAMETHYLBUTYL)PHENOXY]ETHOXY}ETHANOL, 4-BROMO-3-(5'-CARBOXY-4'-CHLORO-2'-FLUOROPHENYL)-1-METHYL-5-TRIFLUOROMETHYL-PYRAZOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Koch, M, Breithaupt, C, Kiefersauer, R, Freigang, J, Huber, R, Messerschmidt, A. | | Deposit date: | 2004-02-19 | | Release date: | 2004-04-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of protoporphyrinogen IX oxidase: a key enzyme in haem and chlorophyll biosynthesis.
Embo J., 23, 2004
|
|
3CJJ
 
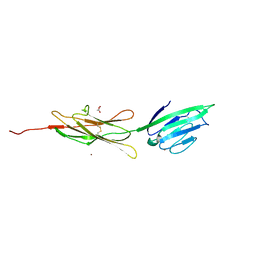 | | Crystal structure of human rage ligand-binding domain | | Descriptor: | ACETATE ION, Advanced glycosylation end product-specific receptor, ZINC ION | | Authors: | Koch, M, Dattilo, B.M, Schiefner, A, Diez, J, Chazin, W.J, Fritz, G. | | Deposit date: | 2008-03-13 | | Release date: | 2009-03-24 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for ligand recognition and activation of RAGE.
Structure, 18, 2010
|
|
4DUQ
 
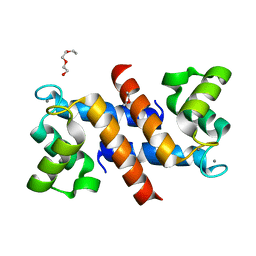 | |
1ZT4
 
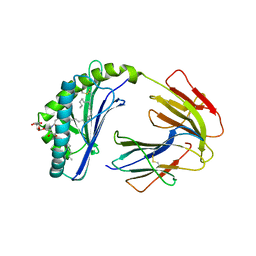 | | The crystal structure of human CD1d with and without alpha-Galactosylceramide | | Descriptor: | Beta-2-microglobulin, N-{(1S,2R,3S)-1-[(ALPHA-D-GALACTOPYRANOSYLOXY)METHYL]-2,3-DIHYDROXYHEPTADECYL}HEXACOSANAMIDE, T-cell surface glycoprotein CD1d | | Authors: | Koch, M, Stronge, V.S, Shepherd, D, Gadola, S.D, Mathew, B, Ritter, G, Fersht, A.R, Besra, G.S, Schmidt, R.R, Jones, E.Y, Cerundolo, V, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-05-26 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of human CD1d with and without alpha-galactosylceramide
Nat.Immunol., 6, 2005
|
|
3BEW
 
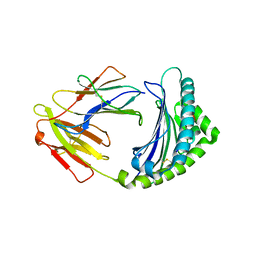 | | 10mer Crystal Structure of chicken MHC class I haplotype B21 | | Descriptor: | 10-mer from Tubulin beta-6 chain, Beta-2-microglobulin, Major histocompatibility complex class I glycoprotein haplotype B21 | | Authors: | Koch, M, Camp, S, Collen, T, Avila, D, Salomonsen, J, Wallny, H.J, van Hateren, A, Hunt, L, Jacob, J.P, Johnston, F, Marston, D.A, Shaw, I, Dunbar, P.R, Cerundolo, V, Jones, E.Y, Kaufman, J. | | Deposit date: | 2007-11-20 | | Release date: | 2008-01-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of an MHC class I molecule from b21 chickens illustrate promiscuous Peptide binding
Immunity, 27, 2007
|
|
2RGI
 
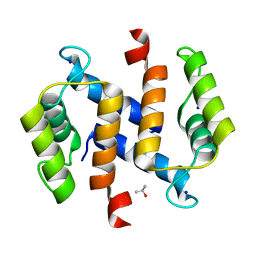 | |
5ZNF
 
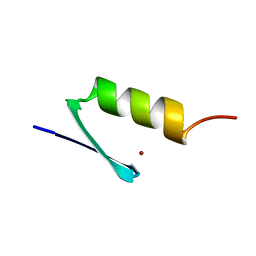 | |
7ZNF
 
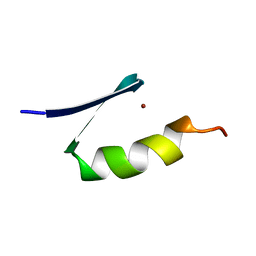 | |
2XPL
 
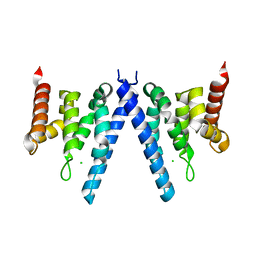 | | Crystal structure of Iws1(Spn1) conserved domain from Encephalitozoon cuniculi | | Descriptor: | CHLORIDE ION, IWS1 | | Authors: | Koch, M, Diebold, M.-L, Cura, V, Cavarelli, J, Romier, C. | | Deposit date: | 2010-08-27 | | Release date: | 2010-11-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Structure of an Iws1/Spt6 Complex Reveals an Interaction Domain Conserved in Tfiis, Elongin a and Med26
Embo J., 29, 2010
|
|
1AUU
 
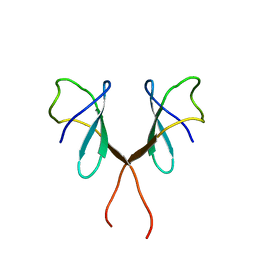 | |
2KKC
 
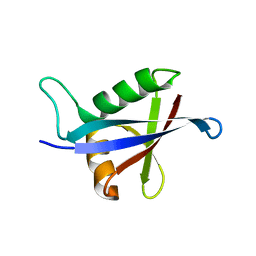 | | NMR structure of the p62 PB1 domain | | Descriptor: | Sequestosome-1 | | Authors: | Yokochi, M, Inagaki, F. | | Deposit date: | 2009-06-18 | | Release date: | 2009-09-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the p62 PB1 domain, a key protein in autophagy and NF-kappaB signaling pathway
J.Biomol.Nmr, 45, 2009
|
|
9D77
 
 | | Crystal form of Netrin-1 mimics nanotubes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Meier, M, Krahn, N.J, McDougall, M.D, Rafiei, F, Koch, M, Stetefeld, J. | | Deposit date: | 2024-08-16 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Mechanistic insights in the higher-order protein assemblies of netrin-1
To Be Published
|
|
4WNX
 
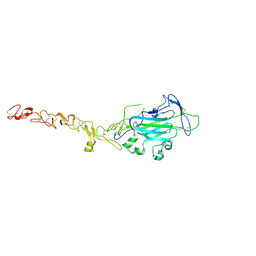 | | Netrin 4 lacking the C-terminal Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | McDougall, M, Patel, T, Reuten, R, Meier, M, Koch, M, Stetefeld, J. | | Deposit date: | 2014-10-14 | | Release date: | 2016-02-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.723 Å) | | Cite: | Structural decoding of netrin-4 reveals a regulatory function towards mature basement membranes.
Nat Commun, 7, 2016
|
|
4OVE
 
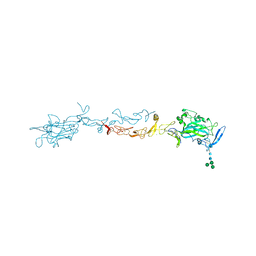 | | X-ray Crystal Structure of Mouse Netrin-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Meier, M, Nikodemus, D, Reuten, R, Patel, T.R, Orriss, G, Okun, N, Koch, M, Stetefeld, J. | | Deposit date: | 2014-02-21 | | Release date: | 2015-02-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural Decoding of the Netrin-1/UNC5 Interaction and its Therapeutical Implications in Cancers.
Cancer Cell, 29, 2016
|
|
2N59
 
 | | Solution Structure of R. palustris CsgH | | Descriptor: | Putative uncharacterized protein CsgH | | Authors: | Hawthorne, W.J, Taylor, J.D, Escalera-Maurer, A, Lambert, S, Koch, M, Scull, N, Sefer, L, Xu, Y, Matthews, S.J. | | Deposit date: | 2015-07-13 | | Release date: | 2016-05-11 | | Method: | SOLUTION NMR | | Cite: | Electrostatically-guided inhibition of Curli amyloid nucleation by the CsgC-like family of chaperones.
Sci Rep, 6, 2016
|
|
6HG7
 
 | | Crystal structure of a collagen II fragment containing the binding site of PEDF and COMP, (POG)4-LKG HRG FTG LQG-POG(4) | | Descriptor: | Collagen alpha-1(II) chain, SULFATE ION | | Authors: | Gebauer, J.M, Koehler, A, Dietmar, H, Gompert, M, Neundorf, I, Zaucke, F, Koch, M, Baumann, U. | | Deposit date: | 2018-08-22 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | COMP and TSP-4 interact specifically with the novel GXKGHR motif only found in fibrillar collagens.
Sci Rep, 8, 2018
|
|
2XP1
 
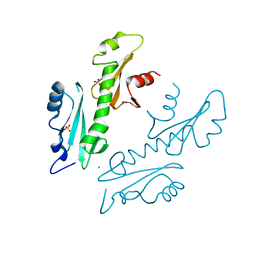 | | Structure of the tandem SH2 domains from Antonospora locustae transcription elongation factor Spt6 | | Descriptor: | CHLORIDE ION, SPT6, SULFATE ION | | Authors: | Diebold, M.-L, Koch, M, Cavarelli, J, Romier, C. | | Deposit date: | 2010-08-24 | | Release date: | 2010-09-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Noncanonical Tandem Sh2 Enables Interaction of Elongation Factor Spt6 with RNA Polymerase II.
J.Biol.Chem., 285, 2010
|
|
2CDG
 
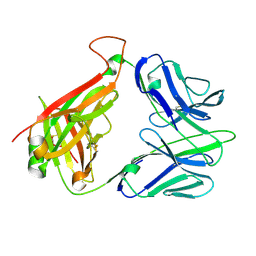 | | Structure and binding kinetics of three different human CD1d-alpha- Galactosylceramide-specific T cell receptors (TCR 5B) | | Descriptor: | TCR 5E | | Authors: | Gadola, S.D, Koch, M, Marles-Wright, J, Lissin, N.M, Sheperd, D, Matulis, G, Harlos, K, Villiger, P.M, Stuart, D.I, Jakobsen, B.K, Cerundolo, V, Jones, E.Y. | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structrue and Binding Kinetics of Three Different Human Cd1D-Alpha-Galactosylceramide-Specific T Cell Receptors
J.Exp.Med., 203, 2006
|
|
2CDE
 
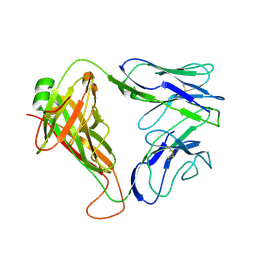 | | Structure and binding kinetics of three different human CD1d-alpha- Galactosylceramide specific T cell receptors - iNKT-TCR | | Descriptor: | INKT-TCR | | Authors: | Gadola, S.D, Koch, M, Marles-Wright, J, Lissin, N.M, Sheperd, D, Matulis, G, Harlos, K, Villiger, P.M, Stuart, D.I, Jakobsen, B.K, Cerundolo, V, Jones, E.Y. | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structrue and Binding Kinetics of Three Different Human Cd1D-Alpha-Galactosylceramide-Specific T Cell Receptors
J.Exp.Med., 203, 2006
|
|
