6YNC
 
 | | Crystal structure of cAMP-dependent Protein Kinase (PKA) in complex with the methylated Fasudil-derived fragment N-methylisoquinoline-5-sulfonamide (soaked) | | Descriptor: | cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha, ~{N}-methylisoquinoline-5-sulfonamide | | Authors: | Oebbeke, M, Wienen-Schmidt, B, Heine, A, Klebe, G. | | Deposit date: | 2020-04-13 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Two Methods, One Goal: Structural Differences between Cocrystallization and Crystal Soaking to Discover Ligand Binding Poses.
Chemmedchem, 16, 2021
|
|
6YGL
 
 | |
6YGX
 
 | | tRNA-guanine Transglycosylase (TGT) labeled with 5-fluorotryptophan in co-crystallized complex with 6-amino-2-(methylamino)-4-(2-((2R,3R,4S,5R,6S)-3,4,5,6-tetramethoxytetrahydro-2H-pyran-2-yl)ethyl)-1H-imidazo[4,5-g]quinazolin-8(7H)-one | | Descriptor: | 6-azanyl-2-(methylamino)-4-[2-[(2~{R},3~{R},4~{S},5~{R},6~{S})-3,4,5,6-tetramethoxyoxan-2-yl]ethyl]-1,7-dihydroimidazo[4,5-g]quinazolin-8-one, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Nguyen, A, Heine, A, Klebe, G. | | Deposit date: | 2020-03-27 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Co-crystallization, nanoESI-MS and 19F NMR reveal dimer disturbing inhibitors and conformational changes at dimer contacts
To Be Published
|
|
6YQI
 
 | | Crystal structure of cAMP-dependent Protein Kinase (PKA) in complex with long-chain Fasudil-derivative N-[2-(propylamino)ethyl]isoquinoline-5-sulfonamide (soaked) | | Descriptor: | cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha, ~{N}-[2-(propylamino)ethyl]isoquinoline-5-sulfonamide | | Authors: | Oebbeke, M, Wienen-Schmidt, B, Heine, A, Klebe, G. | | Deposit date: | 2020-04-17 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Two Methods, One Goal: Structural Differences between Cocrystallization and Crystal Soaking to Discover Ligand Binding Poses.
Chemmedchem, 16, 2021
|
|
5MB7
 
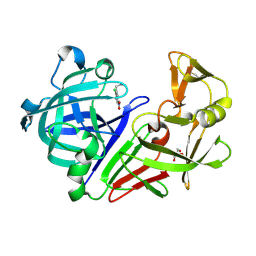 | |
5JS6
 
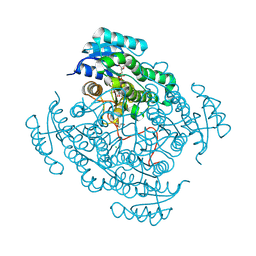 | | Crystal structure of 17beta-hydroxysteroid dehydrogenase 14 T205 variant in complex with NAD. | | Descriptor: | 17-beta-hydroxysteroid dehydrogenase 14, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Bertoletti, N, Marchais-Oberwinkler, S, Heine, A, Klebe, G. | | Deposit date: | 2016-05-07 | | Release date: | 2016-07-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | New Insights into Human 17 beta-Hydroxysteroid Dehydrogenase Type 14: First Crystal Structures in Complex with a Steroidal Ligand and with a Potent Nonsteroidal Inhibitor.
J.Med.Chem., 59, 2016
|
|
5JSF
 
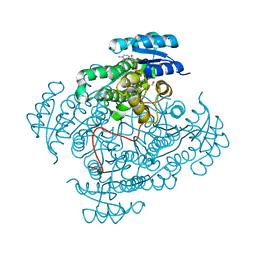 | | Crystal structure of 17beta-hydroxysteroid dehydrogenase 14 S205 variant in complex with NAD. | | Descriptor: | 17-beta-hydroxysteroid dehydrogenase 14, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Bertoletti, N, Marchais-Oberwinkler, S, Heine, A, Klebe, G. | | Deposit date: | 2016-05-08 | | Release date: | 2016-07-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.842 Å) | | Cite: | New Insights into Human 17 beta-Hydroxysteroid Dehydrogenase Type 14: First Crystal Structures in Complex with a Steroidal Ligand and with a Potent Nonsteroidal Inhibitor.
J.Med.Chem., 59, 2016
|
|
5L7T
 
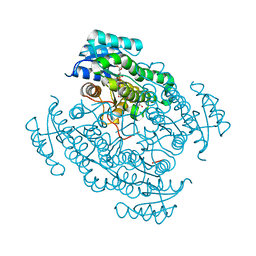 | | 17beta-hydroxysteroid dehydrogenase 14 variant T205 in complex with a non-steroidal inhibitor. | | Descriptor: | (4-fluoranyl-3-oxidanyl-phenyl)-[6-(3-methyl-4-oxidanyl-phenyl)pyridin-2-yl]methanone, 17-beta-hydroxysteroid dehydrogenase 14, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Bertoletti, N, Braun, F, Marchais-Oberwinkler, S, Heine, A, Klebe, G. | | Deposit date: | 2016-06-03 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.983 Å) | | Cite: | First Structure-Activity Relationship of 17 beta-Hydroxysteroid Dehydrogenase Type 14 Nonsteroidal Inhibitors and Crystal Structures in Complex with the Enzyme.
J. Med. Chem., 59, 2016
|
|
5L7Y
 
 | | 17beta-hydroxysteroid dehydrogenase 14 variant T205 in complex with a non-steroidal inhibitor. | | Descriptor: | (4-fluoranyl-3-oxidanyl-phenyl)-[6-(2-fluoranyl-3-oxidanyl-phenyl)pyridin-2-yl]methanone, 17-beta-hydroxysteroid dehydrogenase 14, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Bertoletti, N, Braun, F, Marchais-Oberwinkler, S, Heine, A, Klebe, G. | | Deposit date: | 2016-06-04 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.912 Å) | | Cite: | First Structure-Activity Relationship of 17 beta-Hydroxysteroid Dehydrogenase Type 14 Nonsteroidal Inhibitors and Crystal Structures in Complex with the Enzyme.
J. Med. Chem., 59, 2016
|
|
5L7W
 
 | | 17beta-hydroxysteroid dehydrogenase 14 variant T205 in complex with a non-steroidal inhibitor. | | Descriptor: | 17-beta-hydroxysteroid dehydrogenase 14, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Bertoletti, N, Braun, F, Marchais-Oberwinkler, S, Heine, A, Klebe, G. | | Deposit date: | 2016-06-04 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | First Structure-Activity Relationship of 17 beta-Hydroxysteroid Dehydrogenase Type 14 Nonsteroidal Inhibitors and Crystal Structures in Complex with the Enzyme.
J. Med. Chem., 59, 2016
|
|
5LCP
 
 | |
5LCT
 
 | |
5LCU
 
 | |
5LCQ
 
 | |
5LCR
 
 | |
2Z1W
 
 | | tRNA guanine transglycosylase TGT E235Q mutant in complex with BDI (2-BUTYL-5,6-DIHYDRO-1H-IMIDAZO[4,5-D]PYRIDAZINE-4,7-DIONE) | | Descriptor: | 2-BUTYL-5,6-DIHYDRO-1H-IMIDAZO[4,5-D]PYRIDAZINE-4,7-DIONE, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Tidten, N, Stengl, B, Heine, A, Garcia, G.A, Klebe, G, Reuter, K. | | Deposit date: | 2007-05-16 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Glutamate versus Glutamine Exchange Swaps Substrate Selectivity in tRNA-Guanine Transglycosylase: Insight into the Regulation of Substrate Selectivity by Kinetic and Crystallographic Studies
J.Mol.Biol., 374, 2007
|
|
2Z7K
 
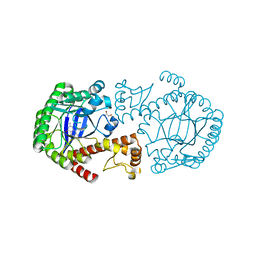 | | tRNA-Guanine transglycosylase (TGT) in complex with 2-Amino-lin-Benzoguanine | | Descriptor: | 2,6-diamino-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Ritschel, T, Heine, A, Klebe, G. | | Deposit date: | 2007-08-24 | | Release date: | 2008-08-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Crystal structure analysis and in silico pKa calculations suggest strong pKa shifts of ligands as driving force for high-affinity binding to TGT
Chembiochem, 10, 2009
|
|
2ZDK
 
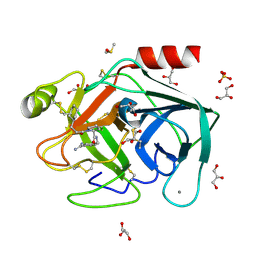 | | Exploring Trypsin S3 Pocket | | Descriptor: | (S)-N-(4-carbamimidoylbenzyl)-1-(3-cyclohexylpropanoyl)pyrrolidine-2-carboxamide, CALCIUM ION, Cationic trypsin, ... | | Authors: | Baum, B, Brandt, T, Heine, A, Klebe, G. | | Deposit date: | 2007-11-26 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Congeneric but still distinct: how closely related trypsin ligands exhibit different thermodynamic and structural properties
J.Mol.Biol., 405, 2011
|
|
2ZDV
 
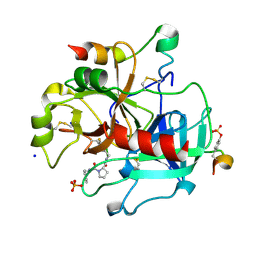 | | Exploring Thrombin S1 pocket | | Descriptor: | D-phenylalanyl-N-(3-fluorobenzyl)-L-prolinamide, Hirudin variant-1, PHOSPHATE ION, ... | | Authors: | Baum, B, Heine, A, Klebe, G. | | Deposit date: | 2007-11-29 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Exploring Thrombin S1 pocket
To be Published
|
|
2ZDM
 
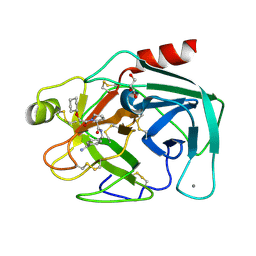 | | Exploring trypsin S3 pocket | | Descriptor: | (S)-N-(4-carbamimidoylbenzyl)-1-(2-(cyclohexyloxy)ethanoyl)pyrrolidine-2-carboxamide, CALCIUM ION, Cationic trypsin, ... | | Authors: | Baum, B, Brandt, T, Heine, A, Klebe, G. | | Deposit date: | 2007-11-26 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Congeneric but still distinct: how closely related trypsin ligands exhibit different thermodynamic and structural properties
J.Mol.Biol., 405, 2011
|
|
2ZHE
 
 | | Exploring thrombin S3 pocket | | Descriptor: | Hirudin variant-2, N-cyclooctylglycyl-N-(4-carbamimidoylbenzyl)-L-prolinamide, SODIUM ION, ... | | Authors: | Brandt, T, Baum, B, Heine, A, Klebe, G. | | Deposit date: | 2008-02-04 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Exploring thrombin S3 pocket
To be Published
|
|
2ZGX
 
 | | Thrombin Inhibition | | Descriptor: | 1-[(2R)-2-aminobutanoyl]-N-(4-carbamimidoylbenzyl)-L-prolinamide, Hirudin variant-1, SODIUM ION, ... | | Authors: | Baum, B, Heine, A, Klebe, G. | | Deposit date: | 2008-01-28 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Think twice: understanding the high potency of bis(phenyl)methane inhibitors of thrombin
J.Mol.Biol., 391, 2009
|
|
2Z1V
 
 | | tRNA guanine transglycosylase E235Q mutant apo structure, pH 8.5 | | Descriptor: | GLYCEROL, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Tidten, N, Stengl, B, Heine, A, Garcia, G.A, Klebe, G, Reuter, K. | | Deposit date: | 2007-05-15 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Glutamate versus Glutamine Exchange Swaps Substrate Selectivity in tRNA-Guanine Transglycosylase: Insight into the Regulation of Substrate Selectivity by Kinetic and Crystallographic Studies
J.Mol.Biol., 374, 2007
|
|
2ZFT
 
 | | Exploring trypsin S3 pocket | | Descriptor: | (S)-N-(4-carbamimidoylbenzyl)-1-(2-(cyclopentylamino)ethanoyl)pyrrolidine-2-carboxamide, CALCIUM ION, Cationic trypsin, ... | | Authors: | Brandt, T, Baum, B, Heine, A, Klebe, G. | | Deposit date: | 2008-01-11 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Congeneric but still distinct: how closely related trypsin ligands exhibit different thermodynamic and structural properties
J.Mol.Biol., 405, 2011
|
|
2ZHF
 
 | | Exploring thrombin S3 pocket | | Descriptor: | (S)-N-(4-carbamimidoylbenzyl)-1-(3-cyclopentylpropanoyl)pyrrolidine-2-carboxamide, GLYCEROL, Hirudin variant-2, ... | | Authors: | Brandt, T, Baum, B, Heine, A, Klebe, G. | | Deposit date: | 2008-02-04 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Exploring thrombin S3 pocket
To be Published
|
|
