6IM3
 
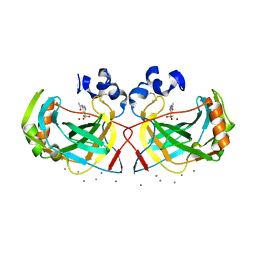 | | Crystal structure of a highly thermostable carbonic anhydrase from Persephonella marina EX-H1 | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, CALCIUM ION, Carbonic anhydrase (Carbonate dehydratase), ... | | Authors: | Jin, M.S, Kim, S, Sung, J, Yeon, J, Choi, S.H. | | Deposit date: | 2018-10-22 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Highly Thermostable alpha-Carbonic Anhydrase from Persephonella marina EX-H1.
Mol.Cells, 42, 2019
|
|
6O6J
 
 | |
6O7C
 
 | |
6O7A
 
 | |
4RDI
 
 | | Crystal structure of E. coli tRNA N6-threonylcarbamoyladenosine dehydratase, TcdA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, tRNA threonylcarbamoyladenosine dehydratase | | Authors: | Park, S.Y, Kim, S, Lee, H. | | Deposit date: | 2014-09-19 | | Release date: | 2015-08-05 | | Last modified: | 2015-10-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Structure of Escherichia coli TcdA (Also Known As CsdL) Reveals a Novel Topology and Provides Insight into the tRNA Binding Surface Required for N(6)-Threonylcarbamoyladenosine Dehydratase Activity.
J.Mol.Biol., 427, 2015
|
|
4RDH
 
 | | Crystal structure of E. coli tRNA N6-threonylcarbamoyladenosine dehydratase, TcdA | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, SULFATE ION, ... | | Authors: | Park, S.Y, Kim, S, Lee, H. | | Deposit date: | 2014-09-19 | | Release date: | 2015-08-05 | | Last modified: | 2015-10-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of Escherichia coli TcdA (Also Known As CsdL) Reveals a Novel Topology and Provides Insight into the tRNA Binding Surface Required for N(6)-Threonylcarbamoyladenosine Dehydratase Activity.
J.Mol.Biol., 427, 2015
|
|
3OU0
 
 | | re-refined 3CS0 | | Descriptor: | Periplasmic serine endoprotease DegP, heptapeptide, pentapeptide | | Authors: | Sauer, R.T, Grant, R.A, Kim, S. | | Deposit date: | 2010-09-14 | | Release date: | 2011-01-19 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Covalent Linkage of Distinct Substrate Degrons Controls Assembly and Disassembly of DegP Proteolytic Cages.
Cell(Cambridge,Mass.), 145, 2011
|
|
2MLP
 
 | | MICROCIN LEADER PEPTIDE FROM E. COLI, NMR, 25 STRUCTURES | | Descriptor: | MCBA PROPEPTIDE | | Authors: | Kim, S, Sinha Roy, R, Walsh, C.T, Baleja, J.D. | | Deposit date: | 1998-01-21 | | Release date: | 1998-07-22 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Role of the microcin B17 propeptide in substrate recognition: solution structure and mutational analysis of McbA1-26.
Chem.Biol., 5, 1998
|
|
7XGF
 
 | |
7XGE
 
 | |
7XGG
 
 | |
1EUJ
 
 | | A NOVEL ANTI-TUMOR CYTOKINE CONTAINS A RNA-BINDING MOTIF PRESENT IN AMINOACYL-TRNA SYNTHETASES | | Descriptor: | ENDOTHELIAL MONOCYTE ACTIVATING POLYPEPTIDE 2 | | Authors: | Kim, Y, Shin, J, Li, R, Cheong, C, Kim, S. | | Deposit date: | 2000-04-17 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel anti-tumor cytokine contains an RNA binding motif present in aminoacyl-tRNA synthetases.
J.Biol.Chem., 275, 2000
|
|
5JJH
 
 | |
6J10
 
 | | Ciclopirox inhibits Hepatitis B Virus secretion by blocking capsid assembly | | Descriptor: | 6-cyclohexyl-4-methyl-1-oxidanyl-pyridin-2-one, Capsid protein | | Authors: | Park, S, Jin, M.S, Cho, Y, Kang, J, Kim, S, Park, M, Park, H, Kim, J, Park, S, Hwang, J, Kim, Y, Kim, Y.J. | | Deposit date: | 2018-12-27 | | Release date: | 2019-04-17 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ciclopirox inhibits Hepatitis B Virus secretion by blocking capsid assembly.
Nat Commun, 10, 2019
|
|
1FYJ
 
 | | SOLUTION STRUCTURE OF MULTI-FUNCTIONAL PEPTIDE MOTIF-1 PRESENT IN HUMAN GLUTAMYL-PROLYL TRNA SYNTHETASE (EPRS). | | Descriptor: | MULTIFUNCTIONAL AMINOACYL-TRNA SYNTHETASE | | Authors: | Jeong, E.-J, Hwang, G.-S, Kim, K.H, Kim, M.J, Kim, S, Kim, K.-S. | | Deposit date: | 2000-10-02 | | Release date: | 2001-03-14 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of multifunctional peptide motifs in human bifunctional tRNA synthetase: identification of RNA-binding residues and functional implications for tandem repeats.
Biochemistry, 39, 2000
|
|
7CPZ
 
 | | Crystal structure of Streptoavidin-C1 from Streptomyces cinamonensis | | Descriptor: | BIOTIN, Mature Streptoavidin-C1 | | Authors: | Jeon, B.J, Kim, S, Lee, J.-H, Kim, M.S, Hwang, K.Y. | | Deposit date: | 2020-08-08 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the structure of mature streptavidin C1 from Streptomyces cinnamonensis reveal the self-binding of the extension C-terminal peptide to biotin-binding sites.
Iucrj, 8, 2021
|
|
7CQ0
 
 | | Crystal structure of Streptoavidin-C1 from Streptomyces cinamonensis | | Descriptor: | Mature Streptoavidin-C1 | | Authors: | Jeon, B.J, Kim, S, Lee, J.-H, Kim, M.S, Hwang, K.Y. | | Deposit date: | 2020-08-08 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Insights into the structure of mature streptavidin C1 from Streptomyces cinnamonensis reveal the self-binding of the extension C-terminal peptide to biotin-binding sites.
Iucrj, 8, 2021
|
|
2UZ8
 
 | | The crystal structure of p18, human translation elongation factor 1 epsilon 1 | | Descriptor: | EUKARYOTIC TRANSLATION ELONGATION FACTOR 1 EPSILON-1, GLYCEROL | | Authors: | Kang, B.S, Kim, K.J, Kim, M.H, Oh, Y.S, Kim, S. | | Deposit date: | 2007-04-26 | | Release date: | 2008-03-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Determination of Three-Dimensional Structure and Residues of the Novel Tumor Suppressor Aimp3/P18 Required for the Interaction with Atm.
J.Biol.Chem., 283, 2008
|
|
6LAI
 
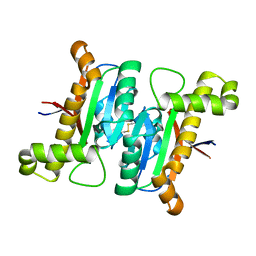 | | The structural basis of the beta-carbonic anhydrase CafD (E54A mutant) of the filamentous fungus Aspergillus fumigatus | | Descriptor: | Carbonic anhydrase, ZINC ION | | Authors: | Jin, M.S, Kim, S, Kim, N.J, Hong, S, Kim, S, Sung, J. | | Deposit date: | 2019-11-12 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structural analysis of non-catalytic zinc binding site in the minor beta-carbonic anhydrase CafD
To be published
|
|
6LAC
 
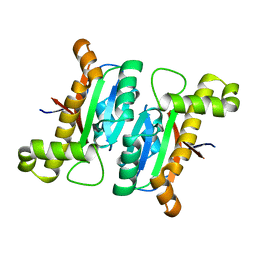 | | The structural basis of the beta-carbonic anhydrase CafD (C39A mutant) of the filamentous fungus Aspergillus fumigatus | | Descriptor: | Carbonic anhydrase, ZINC ION | | Authors: | Jiin, M.S, Kim, S, Kim, N.J, Hong, S, Kim, S, Sung, J. | | Deposit date: | 2019-11-12 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structural analysis of non-catalytic zinc binding site in the minor beta-carbonic anhydrase CafD
To be published
|
|
5B68
 
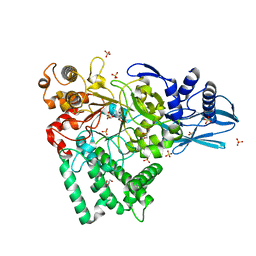 | | Crystal structure of apo amylomaltase from Corynebacterium glutamicum | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-alpha-glucanotransferase, GLYCEROL, ... | | Authors: | Joo, S, Kim, S, Kim, K.-J. | | Deposit date: | 2016-05-25 | | Release date: | 2016-08-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Amylomaltase from Corynebacterium glutamicum.
J.Agric.Food Chem., 64, 2016
|
|
1I69
 
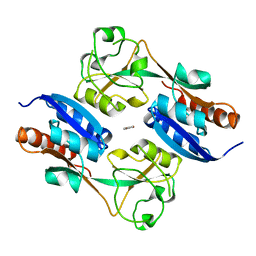 | | CRYSTAL STRUCTURE OF THE REDUCED FORM OF OXYR | | Descriptor: | BENZOIC ACID, HYDROGEN PEROXIDE-INDUCIBLE GENES ACTIVATOR | | Authors: | Choi, H, Kim, S, Ryu, S. | | Deposit date: | 2001-03-02 | | Release date: | 2001-09-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the redox switch in the OxyR transcription factor.
Cell(Cambridge,Mass.), 105, 2001
|
|
1I6A
 
 | | CRYSTAL STRUCTURE OF THE OXIDIZED FORM OF OXYR | | Descriptor: | HYDROGEN PEROXIDE-INDUCIBLE GENES ACTIVATOR | | Authors: | Choi, H, Kim, S, Ryu, S. | | Deposit date: | 2001-03-02 | | Release date: | 2001-09-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of the redox switch in the OxyR transcription factor.
Cell(Cambridge,Mass.), 105, 2001
|
|
6K65
 
 | | Application of anti-helix antibodies in protein structure determination (9014-1P4B) | | Descriptor: | 1P4B variable heavy chain, 1P4B variable light chain, Immunoglobulin G-binding protein A | | Authors: | Lee, J.O, Jin, M.S, Kim, J.W, Kim, S, Lee, H, Cho, G.Y. | | Deposit date: | 2019-06-01 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Application of antihelix antibodies in protein structure determination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6K6B
 
 | | Application of anti-helix antibodies in protein structure determination (8496-3LRH) | | Descriptor: | 3LRH intrabody, Protein A | | Authors: | Lee, J.O, Jin, M.S, Kim, J.W, Kim, S, Lee, H, Cho, G.Y. | | Deposit date: | 2019-06-02 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Application of antihelix antibodies in protein structure determination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
