1RCH
 
 | | SOLUTION NMR STRUCTURE OF RIBONUCLEASE HI FROM ESCHERICHIA COLI, 8 STRUCTURES | | 分子名称: | RIBONUCLEASE HI | | 著者 | Yamazaki, T, Fujiwara, M, Kato, T, Yamasaki, K, Nagayama, K. | | 登録日 | 1995-06-23 | | 公開日 | 1997-02-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of Ribonuclease Hi from Escherichia Coli
Biol.Pharm.Bull., 23, 2000
|
|
6JNA
 
 | | Cryo-EM structure of glutamate dehydrogenase from Thermococcus profundus | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutamate dehydrogenase | | 著者 | Oide, M, Kato, T, Oroguchi, T, Nakasako, M. | | 登録日 | 2019-03-14 | | 公開日 | 2020-02-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Energy landscape of domain motion in glutamate dehydrogenase deduced from cryo-electron microscopy.
Febs J., 287, 2020
|
|
6JN9
 
 | | Cryo-EM structure of glutamate dehydrogenase from Thermococcus profundus | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutamate dehydrogenase | | 著者 | Oide, M, Kato, T, Oroguchi, T, Nakasako, M. | | 登録日 | 2019-03-14 | | 公開日 | 2020-02-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Energy landscape of domain motion in glutamate dehydrogenase deduced from cryo-electron microscopy.
Febs J., 287, 2020
|
|
6JNC
 
 | | Cryo-EM structure of glutamate dehydrogenase from Thermococcus profundus | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutamate dehydrogenase | | 著者 | Oide, M, Kato, T, Oroguchi, T, Nakasako, M. | | 登録日 | 2019-03-14 | | 公開日 | 2020-02-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Energy landscape of domain motion in glutamate dehydrogenase deduced from cryo-electron microscopy.
Febs J., 287, 2020
|
|
6JND
 
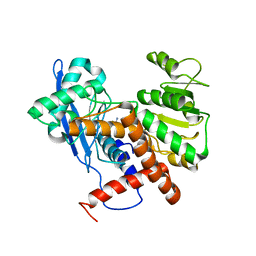 | | Cryo-EM structure of glutamate dehydrogenase from Thermococcus profundus | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutamate dehydrogenase | | 著者 | Oide, M, Kato, T, Oroguchi, T, Nakasako, M. | | 登録日 | 2019-03-14 | | 公開日 | 2020-02-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Energy landscape of domain motion in glutamate dehydrogenase deduced from cryo-electron microscopy.
Febs J., 287, 2020
|
|
6O97
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with propylamycin and bound to mRNA and A-, P-, and E-site tRNAs at 2.75A resolution | | 分子名称: | (1R,2R,3S,4R,6S)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-beta-D-ribofuranosyl]oxy}-3-hydroxyc yclohexyl 2-amino-2,4-dideoxy-4-propyl-alpha-D-glucopyranoside, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | 著者 | Matsushita, T, Sati, G.C, Kondasinghe, N, Pirrone, M.G, Kato, T, Waduge, P, Kumar, H.S, Sanchon, A.C, Dobosz-Bartoszek, M, Shcherbakov, D, Juhas, M, Hobbie, S.N, Schrepfer, T, Chow, C.S, Polikanov, Y.S, Schacht, J, Vasella, A, Bottger, E.C, Crich, D. | | 登録日 | 2019-03-13 | | 公開日 | 2019-04-17 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Design, Multigram Synthesis, and in Vitro and in Vivo Evaluation of Propylamycin: A Semisynthetic 4,5-Deoxystreptamine Class Aminoglycoside for the Treatment of Drug-Resistant Enterobacteriaceae and Other Gram-Negative Pathogens.
J. Am. Chem. Soc., 141, 2019
|
|
8GQY
 
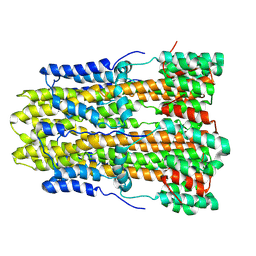 | | CryoEM structure of pentameric MotA from Aquifex aeolicus | | 分子名称: | Motility protein A | | 著者 | Nishikino, T, Takekawa, N, Kishikawa, J, Hirose, M, Onoe, S, Kato, T, Imada, K. | | 登録日 | 2022-08-31 | | 公開日 | 2022-10-26 | | 最終更新日 | 2022-11-09 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure of MotA, a flagellar stator protein, from hyperthermophile.
Biochem.Biophys.Res.Commun., 631, 2022
|
|
8HIQ
 
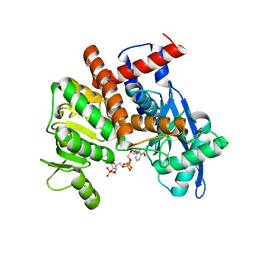 | | cryoEM structure of glutamate dehydrogenase from Thermococcus profundus in complex with NADP | | 分子名称: | Glutamate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Wakabayashi, T, Oide, M, Kato, T, Nakasako, M. | | 登録日 | 2022-11-21 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Coenzyme-binding pathway on glutamate dehydrogenase suggested from multiple-binding sites visualized by cryo-electron microscopy.
Febs J., 290, 2023
|
|
8HJ9
 
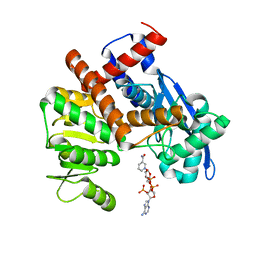 | | cryoEM structure of glutamate dehydrogenase from Thermococcus profundus in complex with NADP | | 分子名称: | Glutamate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Wakabayashi, T, Oide, M, Kato, T, Nakasako, M. | | 登録日 | 2022-11-22 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.12 Å) | | 主引用文献 | Coenzyme-binding pathway on glutamate dehydrogenase suggested from multiple-binding sites visualized by cryo-electron microscopy.
Febs J., 290, 2023
|
|
8HIZ
 
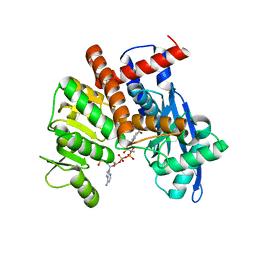 | | cryoEM structure of glutamate dehydrogenase from Thermococcus profundus in complex with NADP | | 分子名称: | Glutamate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Wakabayashi, T, Oide, M, Kato, T, Nakasako, M. | | 登録日 | 2022-11-22 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.08 Å) | | 主引用文献 | Coenzyme-binding pathway on glutamate dehydrogenase suggested from multiple-binding sites visualized by cryo-electron microscopy.
Febs J., 290, 2023
|
|
8HHO
 
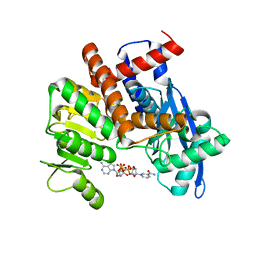 | | cryoEM structure of glutamate dehydrogenase from Thermococcus profundus in complex with NADP | | 分子名称: | Glutamate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Wakabayashi, T, Oide, M, Kato, T, Nakasako, M. | | 登録日 | 2022-11-16 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Coenzyme-binding pathway on glutamate dehydrogenase suggested from multiple-binding sites visualized by cryo-electron microscopy.
Febs J., 290, 2023
|
|
8HJ3
 
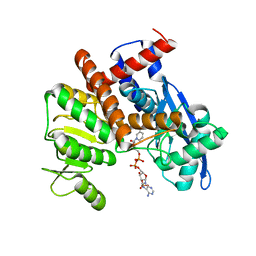 | | cryoEM structure of glutamate dehydrogenase from Thermococcus profundus in complex with NADP | | 分子名称: | Glutamate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Wakabayashi, T, Oide, M, Kato, T, Nakasako, M. | | 登録日 | 2022-11-22 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Coenzyme-binding pathway on glutamate dehydrogenase suggested from multiple-binding sites visualized by cryo-electron microscopy.
Febs J., 290, 2023
|
|
8P6A
 
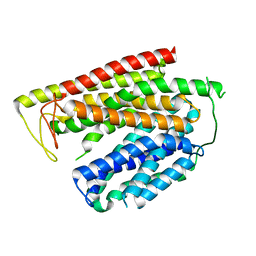 | |
8OMU
 
 | |
6KFK
 
 | |
6LKT
 
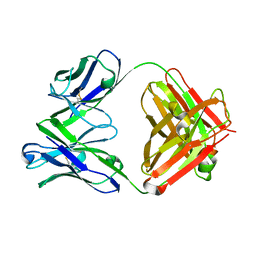 | | Crystal structure of the Fab fragment of murine monoclonal antibody KH-1 against Human herpesvirus 6B | | 分子名称: | antibody Fab Fragment L-chain, antibody Fab fragment H chain | | 著者 | Nishimura, M, Novita, B.D, Kato, T, Tjan, L.H, Wang, B, Wakata, A, Poetranto, A.L, Kawabata, A, Tang, H, Aoshi, T, Mori, Y. | | 登録日 | 2019-12-20 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for the interaction of human herpesvirus 6B tetrameric glycoprotein complex with the cellular receptor, human CD134.
Plos Pathog., 16, 2020
|
|
6LTG
 
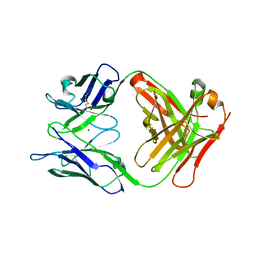 | | Crystal structure of the Fab fragment of murine monoclonal antibody OHV-3 against Human herpesvirus 6B | | 分子名称: | MAGNESIUM ION, antibody Fab fragment H-chain, antibody Fab fragment L-chain | | 著者 | Nishimura, M, Novita, B.D, Kato, T, Tjan, L.H, Wang, B, Wakata, A, Poetranto, A.L, Kawabata, A, Tang, H, Aoshi, T, Mori, Y. | | 登録日 | 2020-01-22 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Structural basis for the interaction of human herpesvirus 6B tetrameric glycoprotein complex with the cellular receptor, human CD134.
Plos Pathog., 16, 2020
|
|
6LU1
 
 | | Cyanobacterial PSI Monomer from T. elongatus by Single Particle CRYO-EM at 3.2 A Resolution | | 分子名称: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | 著者 | Kurisu, G, Coruh, O, Tanaka, H, Gerle, C, Kawamoto, A, Kato, T, Namba, K, Nowaczyk, M.M, Rogner, M, Misumi, Y, Frank, A, Eithar, E.M. | | 登録日 | 2020-01-24 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structure of a functional monomeric Photosystem I from Thermosynechococcus elongatus reveals red chlorophyll cluster.
Commun Biol, 4, 2021
|
|
7VNN
 
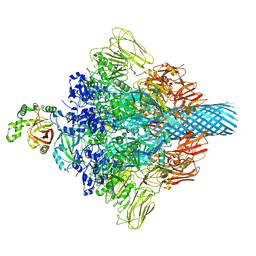 | | Complex structure of Clostridioides difficile enzymatic component (CDTa) and binding component (CDTb) pore with long stem | | 分子名称: | ADP-ribosylating binary toxin binding subunit CdtB, CALCIUM ION, CdtA | | 著者 | Yamada, T, Kawamoto, A, Yoshida, T, Sato, Y, Kato, T, Tsuge, H. | | 登録日 | 2021-10-11 | | 公開日 | 2022-10-26 | | 最終更新日 | 2022-11-02 | | 実験手法 | ELECTRON MICROSCOPY (2.64 Å) | | 主引用文献 | Cryo-EM structures of the translocational binary toxin complex CDTa-bound CDTb-pore from Clostridioides difficile.
Nat Commun, 13, 2022
|
|
7VNJ
 
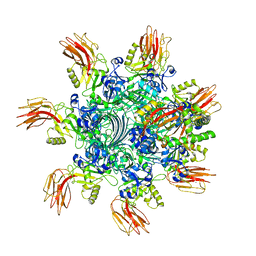 | | Complex structure of Clostridioides difficile enzymatic component (CDTa) and binding component (CDTb) pore with short stem | | 分子名称: | ADP-ribosylating binary toxin binding subunit CdtB, ADP-ribosyltransferase enzymatic component, CALCIUM ION | | 著者 | Yamada, T, Kawamoto, A, Yoshida, T, Sato, Y, Kato, T, Tsuge, H. | | 登録日 | 2021-10-11 | | 公開日 | 2022-10-26 | | 実験手法 | ELECTRON MICROSCOPY (2.56 Å) | | 主引用文献 | Cryo-EM structures of the translocational binary toxin complex CDTa-bound CDTb-pore from Clostridioides difficile.
Nat Commun, 13, 2022
|
|
7WU9
 
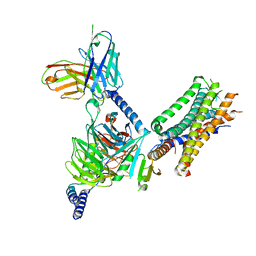 | | Cryo-EM structure of the human EP3-Gi signaling complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Suno, R, Sugita, Y, Morimoto, K, Iwasaki, K, Kato, T, Kobayashi, T. | | 登録日 | 2022-02-07 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (3.375 Å) | | 主引用文献 | Structural insights into the G protein selectivity revealed by the human EP3-G i signaling complex.
Cell Rep, 40, 2022
|
|
7C86
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: Dark state structure | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, RETINAL, ... | | 著者 | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | 登録日 | 2020-05-28 | | 公開日 | 2021-04-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
6LY9
 
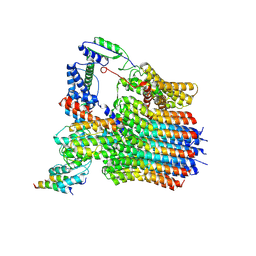 | | The membrane-embedded Vo domain of V/A-ATPase from Thermus thermophilus | | 分子名称: | V-type ATP synthase subunit C, V-type ATP synthase subunit E, V-type ATP synthase subunit I, ... | | 著者 | Kishikawa, J, Nakanishi, A, Furuta, A, Kato, T, Namba, K, Tamakoshi, M, Mitsuoka, K, Yokoyama, K. | | 登録日 | 2020-02-13 | | 公開日 | 2020-09-09 | | 最終更新日 | 2020-09-30 | | 実験手法 | ELECTRON MICROSCOPY (3.93 Å) | | 主引用文献 | Mechanical inhibition of isolated V o from V/A-ATPase for proton conductance.
Elife, 9, 2020
|
|
6LY8
 
 | | V/A-ATPase from Thermus thermophilus, the soluble domain, including V1, d, two EG stalks, and N-terminal domain of a-subunit. | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, V-type ATP synthase alpha chain, V-type ATP synthase beta chain, ... | | 著者 | Kishikawa, J, Nakanishi, A, Furuta, A, Kato, T, Namba, K, Tamakoshi, M, Mitsuoka, K, Yokoyama, K. | | 登録日 | 2020-02-13 | | 公開日 | 2020-09-09 | | 最終更新日 | 2020-09-30 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Mechanical inhibition of isolated V o from V/A-ATPase for proton conductance.
Elife, 9, 2020
|
|
7E6Z
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: 50 microsecond structure | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Archaeal-type opsin 1,Archaeal-type opsin 2, ... | | 著者 | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | 登録日 | 2021-02-24 | | 公開日 | 2021-04-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
