1L1M
 
 | | SOLUTION STRUCTURE OF A DIMER OF LAC REPRESSOR DNA-BINDING DOMAIN COMPLEXED TO ITS NATURAL OPERATOR O1 | | Descriptor: | 5'-D(*AP*AP*AP*TP*TP*GP*TP*TP*AP*TP*CP*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*TP*C)-3', 5'-D(*GP*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*GP*GP*AP*TP*AP*AP*CP*AP*AP*TP*TP*T)-3', Lactose operon repressor | | Authors: | Kalodimos, C.G, Bonvin, A.M.J.J, Salinas, R.K, Wechselberger, R, Boelens, R, Kaptein, R. | | Deposit date: | 2002-02-19 | | Release date: | 2002-06-26 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Plasticity in protein-DNA recognition: lac repressor interacts with its natural operator 01 through alternative conformations of its DNA-binding domain.
EMBO J., 21, 2002
|
|
1OSL
 
 | | Solution structure of a dimeric lactose DNA-binding domain complexed to a nonspecific DNA sequence | | Descriptor: | 5'-D(*CP*GP*AP*TP*AP*AP*GP*AP*TP*AP*TP*CP*TP*TP*AP*TP*CP*G)-3', Lactose operon repressor | | Authors: | Kalodimos, C.G, Bonvin, A.M.J.J, Boelens, R, Kaptein, R. | | Deposit date: | 2003-03-20 | | Release date: | 2004-05-04 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structure and flexibility adaptation in nonspecific and specific protein-DNA complexes.
Science, 305, 2004
|
|
6AMW
 
 | |
6AMV
 
 | |
2L3P
 
 | | Structure of the prolyl cis isomer of the Crk Protein | | Descriptor: | Cis isomer of Crk protein | | Authors: | Kalodimos, C.G, Sarkar, P, Saleh, T, Tzeng, S.R, Birge, R. | | Deposit date: | 2010-09-21 | | Release date: | 2010-12-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for regulation of the Crk signaling protein by a proline switch.
Nat.Chem.Biol., 7, 2011
|
|
2L3S
 
 | | Structure of the autoinhibited Crk | | Descriptor: | Autoinhibited Crk protein | | Authors: | Kalodimos, C.G, Sarkar, P, Saleh, T, Tzeng, S.R, Birge, R. | | Deposit date: | 2010-09-21 | | Release date: | 2010-12-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for regulation of the Crk signaling protein by a proline switch.
Nat.Chem.Biol., 7, 2011
|
|
7MZX
 
 | |
5JTL
 
 | | The structure of chaperone SecB in complex with unstructured proPhoA | | Descriptor: | Alkaline phosphatase, Protein-export protein SecB | | Authors: | Huang, C, Saio, T, Rossi, P, Kalodimos, C.G. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the antifolding activity of a molecular chaperone.
Nature, 537, 2016
|
|
5JTN
 
 | | The structure of chaperone SecB in complex with unstructured proPhoA binding site c | | Descriptor: | Alkaline phosphatase, Protein-export protein SecB | | Authors: | Huang, C, Saio, T, Rossi, P, Kalodimos, C.G. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the antifolding activity of a molecular chaperone.
Nature, 537, 2016
|
|
8FTW
 
 | | FliT-FliJ fusion complex | | Descriptor: | Flagellar FliT protein, Flagellar FliJ protein fusion | | Authors: | Rossi, P, Kalodimos, C.G. | | Deposit date: | 2023-01-13 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Chaperone Recycling in Late-Stage Flagellar Assembly.
J.Mol.Biol., 435, 2023
|
|
8FTX
 
 | | FlgN-FliJ fusion complex | | Descriptor: | Flagella biosynthesis chaperone FlgN, Flagellar FliJ protein fusion | | Authors: | Rossi, P, Kalodimos, C.G. | | Deposit date: | 2023-01-13 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Chaperone Recycling in Late-Stage Flagellar Assembly.
J.Mol.Biol., 435, 2023
|
|
2VDA
 
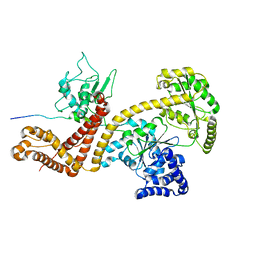 | | Solution structure of the SecA-signal peptide complex | | Descriptor: | MALTOPORIN, TRANSLOCASE SUBUNIT SECA | | Authors: | Gelis, I, Bonvin, A.M.J.J, Keramisanou, D, Koukaki, M, Gouridis, G, Karamanou, S, Economou, A, Kalodimos, C.G. | | Deposit date: | 2007-10-01 | | Release date: | 2007-11-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Signal-Sequence Recognition by the Translocase Motor Seca as Determined by NMR
Cell(Cambridge,Mass.), 131, 2007
|
|
2WC2
 
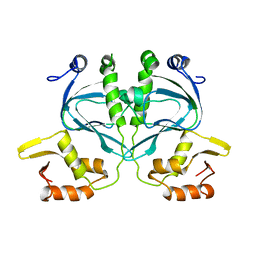 | |
4BHP
 
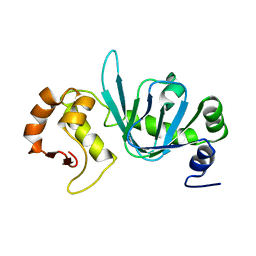 | |
4BH9
 
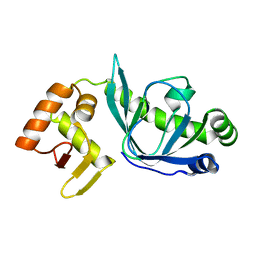 | |
6CH2
 
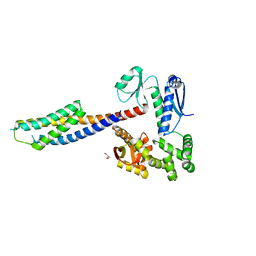 | | Crystal structure of the cytoplasmic domain of FlhA and FliT-FliD complex | | Descriptor: | Flagellar biosynthesis protein FlhA, Flagellar hook-associated protein 2,Flagellar protein FliT, GLYCEROL | | Authors: | Xing, Q, Shi, K, Kalodimos, C.G. | | Deposit date: | 2018-02-21 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of chaperone-substrate complexes docked onto the export gate in a type III secretion system.
Nat Commun, 9, 2018
|
|
6CH3
 
 | |
6D6S
 
 | |
6CH1
 
 | |
7N00
 
 | | Anaplastic lymphoma kinase (ALK) extracellular fragment of ligand binding region 648-1025 in complex with AUG-alpha | | Descriptor: | ALK and LTK ligand 2, ALK tyrosine kinase receptor | | Authors: | Reshetnyak, A.V, Myasnikov, A.G, Rossi, P, Miller, D.J, Kalodimos, C.G. | | Deposit date: | 2021-05-24 | | Release date: | 2021-11-24 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Mechanism for the activation of the anaplastic lymphoma kinase receptor.
Nature, 600, 2021
|
|
7MZY
 
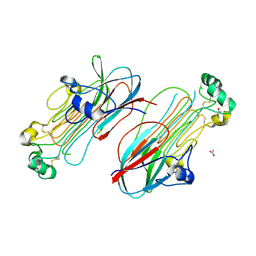 | | Anaplastic lymphoma kinase (ALK) extracellular fragment of ligand binding region 673-986 | | Descriptor: | ACETATE ION, ALK tyrosine kinase receptor | | Authors: | Reshetnyak, A.V, Sowaileh, M, Miller, D.J, Rossi, P, Myasnikov, A.G, Kalodimos, C.G. | | Deposit date: | 2021-05-24 | | Release date: | 2021-11-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanism for the activation of the anaplastic lymphoma kinase receptor.
Nature, 600, 2021
|
|
7MZZ
 
 | |
7MZW
 
 | |
6XRG
 
 | |
6XR6
 
 | |
