7Q1C
 
 | | Crystal structure of Trypanosoma cruzi histone deacetylase DAC2 complexed with a hydroxamate inhibitor | | Descriptor: | (E)-3-dibenzofuran-4-yl-N-oxidanyl-prop-2-enamide, Histone deacetylase DAC2, POTASSIUM ION, ... | | Authors: | Ramos-Morales, E, Marek, M, Romier, C. | | Deposit date: | 2021-10-18 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Species-selective targeting of pathogens revealed by the atypical structure and active site of Trypanosoma cruzi histone deacetylase DAC2.
Cell Rep, 37, 2021
|
|
7Q1B
 
 | | Crystal structure of Trypanosoma cruzi histone deacetylase DAC2 complexed with Quisinostat | | Descriptor: | 2-[4-[[(1-methylindol-3-yl)methylamino]methyl]piperidin-1-yl]-~{N}-oxidanyl-pyrimidine-5-carboxamide, GLYCEROL, Histone deacetylase DAC2, ... | | Authors: | Marek, M, Ramos-Morales, E, Romier, C. | | Deposit date: | 2021-10-18 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Species-selective targeting of pathogens revealed by the atypical structure and active site of Trypanosoma cruzi histone deacetylase DAC2.
Cell Rep, 37, 2021
|
|
7P3S
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 12 | | Descriptor: | GLYCEROL, Histone deacetylase, POTASSIUM ION, ... | | Authors: | Shaik, T.B, Romier, C. | | Deposit date: | 2021-07-08 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.546 Å) | | Cite: | Synthesis, structure-activity relationships, cocrystallization and cellular characterization of novel smHDAC8 inhibitors for the treatment of schistosomiasis.
Eur.J.Med.Chem., 225, 2021
|
|
2VCG
 
 | | Crystal structure of a HDAC-like protein HDAH from Bordetella sp. with the bound inhibitor ST-17 | | Descriptor: | CHLORIDE ION, GLYCEROL, HISTONE DEACETYLASE-LIKE AMIDOHYDROLASE, ... | | Authors: | Dickmanns, A, Strasser, A, Ficner, R. | | Deposit date: | 2007-09-24 | | Release date: | 2008-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phenylalanine-Containing Hydroxamic Acids as Selective Inhibitors of Class Iib Histone Deacetylases (Hdacs).
Bioorg.Med.Chem., 16, 2008
|
|
6QPL
 
 | | Crystal structure of Spindlin1 in complex with the inhibitor MS31 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Johansson, C, Krojer, T, Xiong, Y, Jin, J, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2019-02-14 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of a Potent and Selective Fragment-like Inhibitor of Methyllysine Reader Protein Spindlin 1 (SPIN1).
J.Med.Chem., 62, 2019
|
|
6WJ5
 
 | | Structure of human TRPA1 in complex with inhibitor GDC-0334 | | Descriptor: | (4R,5S)-4-fluoro-1-[(4-fluorophenyl)sulfonyl]-5-methyl-N-({5-(trifluoromethyl)-2-[2-(trifluoromethyl)pyrimidin-5-yl]pyridin-4-yl}methyl)-L-prolinamide, Transient receptor potential cation channel subfamily A member 1 | | Authors: | Rohou, A, Rouge, L, Arthur, C.P, Volgraf, M, Chen, H. | | Deposit date: | 2020-04-11 | | Release date: | 2021-02-17 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A TRPA1 inhibitor suppresses neurogenic inflammation and airway contraction for asthma treatment.
J.Exp.Med., 218, 2021
|
|
7AZN
 
 | | Structure of mouse AsterC (GramD1c) with a new cholesterol-derived compound | | Descriptor: | 20alpha-hydroxy-20-(5-methylhexyl)cholesterol, ETHANOL, GLYCEROL, ... | | Authors: | Romartinez-Alonso, B, Sirvydis, K, Kim, Y, Xiao, X, Jung, M, Tontonoz, P, Schwabe, J. | | Deposit date: | 2020-11-16 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Selective Aster inhibitors distinguish vesicular and nonvesicular sterol transport mechanisms.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6FU1
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a n-alkyl hydroxamate | | Descriptor: | GLYCEROL, Histone deacetylase, POTASSIUM ION, ... | | Authors: | Marek, M, Shaik, T.B, Romier, C. | | Deposit date: | 2018-02-26 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.548 Å) | | Cite: | A Novel Class of Schistosoma mansoni Histone Deacetylase 8 (HDAC8) Inhibitors Identified by Structure-Based Virtual Screening and In Vitro Testing.
Molecules, 23, 2018
|
|
8G1Z
 
 | |
8G20
 
 | |
6ETS
 
 | | Crystal structure of KDM4D with tetrazolhydrazide compound 1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-10-27 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.333 Å) | | Cite: | Structure-Based Screening of Tetrazolylhydrazide Inhibitors versus KDM4 Histone Demethylases.
Chemmedchem, 14, 2019
|
|
6ETT
 
 | | Crystal structure of KDM4D with tetrazole compound 4 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-10-27 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.257 Å) | | Cite: | Structure-Based Screening of Tetrazolylhydrazide Inhibitors versus KDM4 Histone Demethylases.
Chemmedchem, 14, 2019
|
|
6ETV
 
 | | Crystal structure of KDM4D with tetrazolhydrazide compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-10-27 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structure-Based Screening of Tetrazolylhydrazide Inhibitors versus KDM4 Histone Demethylases.
Chemmedchem, 14, 2019
|
|
6ETU
 
 | | Crystal structure of KDM4D with tetrazolhydrazide compound 7 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-10-27 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structure-Based Screening of Tetrazolylhydrazide Inhibitors versus KDM4 Histone Demethylases.
Chemmedchem, 14, 2019
|
|
6ETW
 
 | | Crystal structure of KDM4D with tetrazolhydrazide compound 3 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-10-27 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure-Based Screening of Tetrazolylhydrazide Inhibitors versus KDM4 Histone Demethylases.
Chemmedchem, 14, 2019
|
|
6ETG
 
 | | Crystal structure of KDM4D with tetrazolhydrazide compound 6 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-10-26 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.279 Å) | | Cite: | Structure-Based Screening of Tetrazolylhydrazide Inhibitors versus KDM4 Histone Demethylases.
Chemmedchem, 14, 2019
|
|
6XFP
 
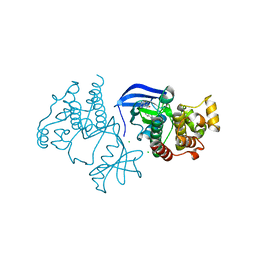 | | Crystal Structure of BRAF kinase domain bound to Belvarafenib | | Descriptor: | 4-amino-N-{1-[(3-chloro-2-fluorophenyl)amino]-6-methylisoquinolin-5-yl}thieno[3,2-d]pyrimidine-7-carboxamide, CHLORIDE ION, Serine/threonine-protein kinase B-raf | | Authors: | Yin, J, Sudhamsu, J. | | Deposit date: | 2020-06-16 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ARAF mutations confer resistance to the RAF inhibitor belvarafenib in melanoma.
Nature, 594, 2021
|
|
7EYC
 
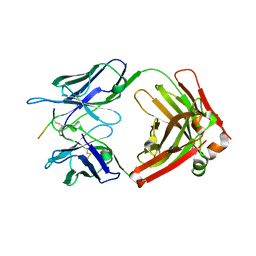 | | Crystal structure of Tau and acetylated tau peptide antigen | | Descriptor: | ACETYLATED TAU PEPTIDE, antibody, Heavy chain, ... | | Authors: | Hong, M, Park, J. | | Deposit date: | 2021-05-30 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Monoclonal antibody Y01 prevents tauopathy progression induced by lysine 280-acetylated tau in cell and mouse models.
J.Clin.Invest., 133, 2023
|
|
6RWJ
 
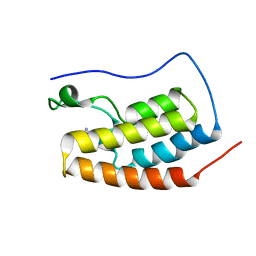 | | Crystal Structure of BRD4(1) bound to inhibitor BUG0 (6) | | Descriptor: | Bromodomain-containing protein 4, ~{N},3-dimethyl-4-oxidanylidene-5,6,7,8-tetrahydro-2~{H}-cyclohepta[c]pyrrole-1-carboxamide | | Authors: | Huegle, M. | | Deposit date: | 2019-06-05 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 4-Acyl Pyrroles as Dual BET-BRD7/9 Bromodomain Inhibitors Address BETi Insensitive Human Cancer Cell Lines.
J.Med.Chem., 63, 2020
|
|
6S4B
 
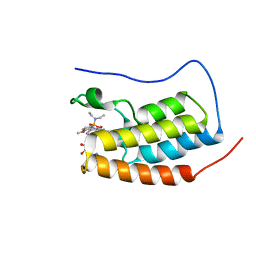 | | Crystal Structure of BRD4(1) bound to inhibitor BUX1 (8) | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Bromodomain-containing protein 4, CALCIUM ION, ... | | Authors: | Huegle, M. | | Deposit date: | 2019-06-27 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 4-Acyl Pyrroles as Dual BET-BRD7/9 Bromodomain Inhibitors Address BETi Insensitive Human Cancer Cell Lines.
J.Med.Chem., 63, 2020
|
|
6S6K
 
 | | Crystal Structure of BRD4(1) bound to inhibitor BUX2 (9) | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Bromodomain-containing protein 4, ~{N}-[5-(azepan-1-ylsulfonyl)-2-methoxy-phenyl]-3-methyl-4-oxidanylidene-5,6,7,8-tetrahydro-2~{H}-cyclohepta[c]pyrrole-1-carboxamide | | Authors: | Huegle, M. | | Deposit date: | 2019-07-03 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 4-Acyl Pyrroles as Dual BET-BRD7/9 Bromodomain Inhibitors Address BETi Insensitive Human Cancer Cell Lines.
J.Med.Chem., 63, 2020
|
|
6SA2
 
 | | Crystal Structure of BRD4(1) bound to inhibitor BUX3 (10) | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-methoxy-5-morpholin-4-ylsulfonyl-phenyl)-3-methyl-4-oxidanylidene-5,6,7,8-tetrahydro-2~{H}-cyclohepta[c]pyrrole-1-carboxamide | | Authors: | Huegle, M. | | Deposit date: | 2019-07-16 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 4-Acyl Pyrroles as Dual BET-BRD7/9 Bromodomain Inhibitors Address BETi Insensitive Human Cancer Cell Lines.
J.Med.Chem., 63, 2020
|
|
6SAH
 
 | | Crystal Structure of BRD4(1) bound to inhibitor BUX5 (11) | | Descriptor: | Bromodomain-containing protein 4, ~{N}-(2-methoxy-5-piperidin-1-ylsulfonyl-phenyl)-3-methyl-4-oxidanylidene-5,6,7,8-tetrahydro-2~{H}-cyclohepta[c]pyrrole-1-carboxamide | | Authors: | Huegle, M. | | Deposit date: | 2019-07-16 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 4-Acyl Pyrroles as Dual BET-BRD7/9 Bromodomain Inhibitors Address BETi Insensitive Human Cancer Cell Lines.
J.Med.Chem., 63, 2020
|
|
6SA3
 
 | | Crystal Structure of BRD4(1) bound to inhibitor BUX4 (13) | | Descriptor: | Bromodomain-containing protein 4, ~{N}-[2-methoxy-5-(4-methylpiperazin-1-yl)sulfonyl-phenyl]-3-methyl-4-oxidanylidene-5,6,7,8-tetrahydro-2~{H}-cyclohepta[c]pyrrole-1-carboxamide | | Authors: | Huegle, M. | | Deposit date: | 2019-07-16 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 4-Acyl Pyrroles as Dual BET-BRD7/9 Bromodomain Inhibitors Address BETi Insensitive Human Cancer Cell Lines.
J.Med.Chem., 63, 2020
|
|
6SB8
 
 | | Crystal Structure of BRD4(1) bound to inhibitor BUX14 (7) | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Bromodomain-containing protein 4, ~{N}-[5-(diethylsulfamoyl)-2-oxidanyl-phenyl]-3-methyl-4-oxidanylidene-5,6,7,8-tetrahydro-2~{H}-cyclohepta[c]pyrrole-1-carboxamide | | Authors: | Huegle, M. | | Deposit date: | 2019-07-19 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 4-Acyl Pyrroles as Dual BET-BRD7/9 Bromodomain Inhibitors Address BETi Insensitive Human Cancer Cell Lines.
J.Med.Chem., 63, 2020
|
|
