8DA3
 
 | | Coevolved affibody-Z domain pair LL1.c1 | | Descriptor: | Affibody LL1.FILF, Immunoglobulin G-binding protein A, MALONATE ION, ... | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
8DA9
 
 | | Coevolved affibody-Z domain pair LL2.c3 | | Descriptor: | Affibody LL2.FIIV, GLYCEROL, Immunoglobulin G-binding protein A, ... | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
8DA4
 
 | | Coevolved affibody-Z domain pair LL1.c2 | | Descriptor: | Affibody LL1.FIVM, Immunoglobulin G-binding protein A, SULFATE ION, ... | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
8DA5
 
 | | Coevolved affibody-Z domain pair LL1.c4 | | Descriptor: | GLYCEROL, Immunoglobulin G-binding protein A, affibody LL1.FIVM | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
8DA6
 
 | | Coevolved affibody-Z domain pair LL1.c5 | | Descriptor: | Affibody LL1.FIIM, Immunoglobulin G-binding protein A | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
8DA8
 
 | | Coevolved affibody-Z domain pair LL2.c1 | | Descriptor: | Affibody LL2.FIIK, GLYCEROL, Immunoglobulin G-binding protein A | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
8DAC
 
 | | Coevolved affibody-Z domain pair LL2.c22 | | Descriptor: | Affibody LL2.FILV, GLYCEROL, Immunoglobulin G-binding protein A | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
8DA7
 
 | | Coevolved affibody-Z domain pair LL1.c6 | | Descriptor: | Immunoglobulin G-binding protein A, MALONATE ION, affibody LL1.FIFV | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
8DAB
 
 | | Coevolved affibody-Z domain pair LL2.c17 | | Descriptor: | Affibody LL2.IVVY, Immunoglobulin G-binding protein A | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.134 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
8DAA
 
 | | Coevolved affibody-Z domain pair LL2.c7 | | Descriptor: | Affibody LL2.FIVK, Immunoglobulin G-binding protein A, MALONATE ION | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
8DZ8
 
 | |
8FFE
 
 | | Crystal structure of LRP6 E1E2 domains bound to YW210.09 Fab and engineered XWnt8 peptide | | Descriptor: | GLYCEROL, Low-density lipoprotein receptor-related protein 6, SODIUM ION, ... | | Authors: | Jude, K.M, Tsutsumi, N, Waghray, D, Garcia, K.C. | | Deposit date: | 2022-12-08 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure of the Wnt-Frizzled-LRP6 initiation complex reveals the basis for coreceptor discrimination.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7N3T
 
 | | TrkA ECD complex with designed miniprotein ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jude, K.M, Cao, L, Garcia, K.C. | | Deposit date: | 2021-06-01 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Design of protein-binding proteins from the target structure alone.
Nature, 605, 2022
|
|
7RAA
 
 | | Designed StabIL-2 seq15 | | Descriptor: | Interleukin-2, MAGNESIUM ION | | Authors: | Jude, K.M, Chu, A.E, Huang, P.-S, Garcia, K.C. | | Deposit date: | 2021-06-30 | | Release date: | 2022-03-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Interleukin-2 superkines by computational design.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RA9
 
 | | Designed StabIL-2 seq1 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Interleukin-2, PHOSPHATE ION | | Authors: | Jude, K.M, Chu, A.E, Huang, P.-S, Garcia, K.C. | | Deposit date: | 2021-06-30 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interleukin-2 superkines by computational design.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6BJ2
 
 | | TCR589 in complex with HIV(Pol448-456)/HLA-B35 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Jude, K.M, Sibener, L.V, Garcia, K.C. | | Deposit date: | 2017-11-03 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Isolation of a Structural Mechanism for Uncoupling T Cell Receptor Signaling from Peptide-MHC Binding.
Cell, 174, 2018
|
|
6BJ3
 
 | | TCR55 in complex with HIV(Pol448-456)/HLA-B35 | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, HIV Pol B35 peptide, ... | | Authors: | Jude, K.M, Sibener, L.V, Garcia, K.C. | | Deposit date: | 2017-11-03 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Isolation of a Structural Mechanism for Uncoupling T Cell Receptor Signaling from Peptide-MHC Binding.
Cell, 174, 2018
|
|
6BJ8
 
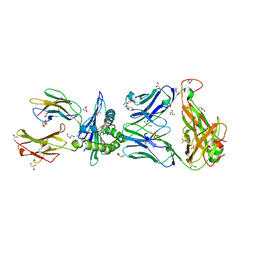 | | TCR55 in complex with Pep20/HLA-B35 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Jude, K.M, Sibener, L.V, Yang, X, Garcia, K.C. | | Deposit date: | 2017-11-05 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Isolation of a Structural Mechanism for Uncoupling T Cell Receptor Signaling from Peptide-MHC Binding.
Cell, 174, 2018
|
|
6WW2
 
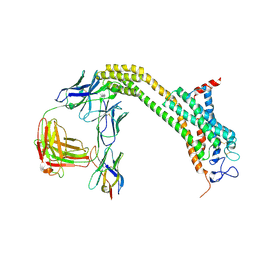 | | Structure of human Frizzled5 by fiducial-assisted cryo-EM | | Descriptor: | Frizzled-5,Soluble cytochrome b562, anti-BRIL Fab Heavy chain, anti-BRIL Fab Light chain, ... | | Authors: | Tsutsumi, N, Jude, K.M, Gati, C, Garcia, K.C. | | Deposit date: | 2020-05-07 | | Release date: | 2020-08-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of human Frizzled5 by fiducial-assisted cryo-EM supports a heterodimeric mechanism of canonical Wnt signaling.
Elife, 9, 2020
|
|
7R86
 
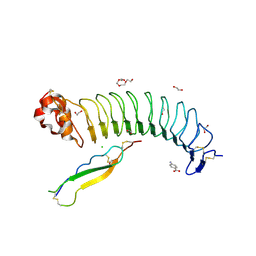 | | Structure of mouse BAI1 (ADGRB1) in complex with mouse Nogo receptor (RTN4R) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-AMINOBENZOIC ACID, ... | | Authors: | Miao, Y, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-06-26 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | RTN4/NoGo-receptor binding to BAI adhesion-GPCRs regulates neuronal development.
Cell, 184, 2021
|
|
7R85
 
 | | Structure of mouse Bai1 (ADGRB1) TSR3 domain | | Descriptor: | Vasculostatin-120, alpha-D-mannopyranose, beta-D-glucopyranose-(1-3)-alpha-L-fucopyranose | | Authors: | Miao, Y, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-06-26 | | Release date: | 2021-11-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | RTN4/NoGo-receptor binding to BAI adhesion-GPCRs regulates neuronal development.
Cell, 184, 2021
|
|
8CTG
 
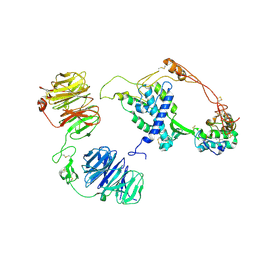 | | Extracellular architecture of an engineered canonical Wnt signaling ternary complex | | Descriptor: | Frizzled-8, Low-density lipoprotein receptor-related protein 6, PALMITOLEIC ACID, ... | | Authors: | Tsutsumi, N, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-05-14 | | Release date: | 2023-03-15 | | Last modified: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Wnt-Frizzled-LRP6 initiation complex reveals the basis for coreceptor discrimination.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8CX4
 
 | | TCR-antigen complex AS8.4-YEIH-HLA*B27 | | Descriptor: | AS8.4a, AS8.4b, Beta-2-microglobulin, ... | | Authors: | Yang, X, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-05-19 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides.
Nature, 612, 2022
|
|
5T5W
 
 | | Structure of an affinity matured lambda-IFN/IFN-lambdaR1/IL-10Rbeta receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interferon lambda receptor 1, Interferon lambda-3, ... | | Authors: | Mendoza, J.L, Jude, K.M, Garcia, K.C. | | Deposit date: | 2016-08-31 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.847 Å) | | Cite: | The IFN-lambda-IFN-lambda R1-IL-10R beta Complex Reveals Structural Features Underlying Type III IFN Functional Plasticity.
Immunity, 46, 2017
|
|
6WDP
 
 | | Interleukin 12 receptor subunit beta-1 | | Descriptor: | GLYCEROL, Interleukin-12 receptor subunit beta-1, SULFATE ION | | Authors: | Spangler, J.B, Thomas, C, Jude, K.M, Garcia, K.C. | | Deposit date: | 2020-04-01 | | Release date: | 2021-02-24 | | Last modified: | 2021-12-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural basis for IL-12 and IL-23 receptor sharing reveals a gateway for shaping actions on T versus NK cells.
Cell, 184, 2021
|
|
