4TLK
 
 | | Crystal Structure of Human Transthyretin Ser85Pro/Glu92Pro Mutant | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SODIUM ION, ... | | Authors: | Saelices, L, Cascio, D, Sawaya, M, Eisenberg, D.S. | | Deposit date: | 2014-05-30 | | Release date: | 2015-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Uncovering the Mechanism of Aggregation of Human Transthyretin.
J.Biol.Chem., 290, 2015
|
|
4TM9
 
 | |
7V29
 
 | | Crystal structure of FGFR4 with a dual-warhead covalent inhhibitor | | Descriptor: | Fibroblast growth factor receptor 4, N-[2-[[3-(3,5-dimethoxyphenyl)-2-oxidanylidene-1-[3-(4-propanoylpiperazin-1-yl)propyl]-4H-pyrimido[4,5-d]pyrimidin-7-yl]amino]phenyl]propanamide, SULFATE ION | | Authors: | Chen, X.J, Jiang, L.Y, Dai, S.Y, Qu, L.Z, Chen, Y.H. | | Deposit date: | 2021-08-07 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.983 Å) | | Cite: | Structure-based design of a dual-warhead covalent inhibitor of FGFR4.
Commun Chem, 5, 2022
|
|
4TJX
 
 | | Crystal structure of protease-associated domain of Arabidopsis VSR1 in complex with aleurain peptide | | Descriptor: | Aleurain peptide, Vacuolar-sorting receptor 1 | | Authors: | Luo, F, Fong, Y.H, Jiang, L.W, Wong, K.B. | | Deposit date: | 2014-05-25 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | How vacuolar sorting receptor proteins interact with their cargo proteins: crystal structures of apo and cargo-bound forms of the protease-associated domain from an Arabidopsis vacuolar sorting receptor.
Plant Cell, 26, 2014
|
|
4TJV
 
 | | Crystal structure of protease-associated domain of Arabidopsis vacuolar sorting receptor 1 | | Descriptor: | IODIDE ION, Vacuolar-sorting receptor 1 | | Authors: | Luo, F, Fong, Y.H, Jiang, L.W, Wong, K.B. | | Deposit date: | 2014-05-25 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | How vacuolar sorting receptor proteins interact with their cargo proteins: crystal structures of apo and cargo-bound forms of the protease-associated domain from an Arabidopsis vacuolar sorting receptor.
Plant Cell, 26, 2014
|
|
8WRZ
 
 | | Cry-EM structure of cannabinoid receptor-arrestin 2 complex | | Descriptor: | (6~{a}~{R},9~{R},10~{a}~{R})-9-(hydroxymethyl)-3-(8-isothiocyanato-2-methyl-octan-2-yl)-6,6-dimethyl-6~{a},7,8,9,10,10~{a}-hexahydrobenzo[c]chromen-1-ol, Beta-arrestin-1, Soluble cytochrome b562,Cannabinoid receptor 1, ... | | Authors: | Wang, Y.X, Wang, T, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2023-10-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of cannabinoid receptor CB1-beta-arrestin complex.
Protein Cell, 15, 2024
|
|
4RIL
 
 | | Structure of the amyloid forming segment, GAVVTGVTAVA, from the NAC domain of Parkinson's disease protein alpha-synuclein, residues 68-78, determined by electron diffraction | | Descriptor: | Alpha-synuclein | | Authors: | Rodriguez, J.A, Ivanova, M, Sawaya, M.R, Cascio, D, Reyes, F, Shi, D, Johnson, L, Guenther, E, Sangwan, S, Hattne, J, Nannenga, B, Brewster, A.S, Messerschmidt, M, Boutet, S, Sauter, N.K, Gonen, T, Eisenberg, D.S. | | Deposit date: | 2014-10-06 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-20 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.43 Å) | | Cite: | Structure of the toxic core of alpha-synuclein from invisible crystals.
Nature, 525, 2015
|
|
4ZS6
 
 | | Receptor binding domain and Fab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S protein, fab Heavy Chain, ... | | Authors: | Yu, X, Wang, X. | | Deposit date: | 2015-05-13 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.166 Å) | | Cite: | Structural basis for the neutralization of MERS-CoV by a human monoclonal antibody MERS-27
Sci Rep, 5, 2015
|
|
5DUP
 
 | | Influenza A virus H5 hemagglutinin globular head in complex with antibody AVFluIgG03 | | Descriptor: | AVFluIgG03 Heavy Chain, AVFluIgG03 Light Chain, Hemagglutinin | | Authors: | Zuo, T, Sun, J, Wang, G, Zhou, P, Wang, X, Zhang, L. | | Deposit date: | 2015-09-20 | | Release date: | 2015-12-16 | | Method: | X-RAY DIFFRACTION (3.052 Å) | | Cite: | Comprehensive analysis of antibody recognition in convalescent humans from highly pathogenic avian influenza H5N1 infection
Nat Commun, 6, 2015
|
|
5DUT
 
 | | Influenza A virus H5 hemagglutinin globular head | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Zuo, T, Sun, J, Wang, G, Zhou, P, Wang, X, Zhang, L. | | Deposit date: | 2015-09-20 | | Release date: | 2015-12-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Comprehensive analysis of antibody recognition in convalescent humans from highly pathogenic avian influenza H5N1 infection
Nat Commun, 6, 2015
|
|
5DUM
 
 | | Crystal structure of influenza A virus H5 hemagglutinin globular head in complex with the Fab of antibody 65C6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 65C6 Heavy Chain, 65C6 Light Chain, ... | | Authors: | Sun, J, Zuo, T, Wang, G, Zhou, P, Zhang, L, Wang, X. | | Deposit date: | 2015-09-19 | | Release date: | 2015-12-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Comprehensive analysis of antibody recognition in convalescent humans from highly pathogenic avian influenza H5N1 infection
Nat Commun, 6, 2015
|
|
5DUR
 
 | | Influenza A virus H5 hemagglutinin globular head in complex with antibody 100F4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of Antibody 100F4, Hemagglutinin, ... | | Authors: | Zuo, T, Sun, J, Wang, G, Zhou, P, Wang, X, Zhang, L. | | Deposit date: | 2015-09-20 | | Release date: | 2015-12-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Comprehensive analysis of antibody recognition in convalescent humans from highly pathogenic avian influenza H5N1 infection
Nat Commun, 6, 2015
|
|
1OQD
 
 | | Crystal structure of sTALL-1 and BCMA | | Descriptor: | Tumor necrosis factor ligand superfamily member 13B, soluble form, Tumor necrosis factor receptor superfamily member 17 | | Authors: | Zhang, G. | | Deposit date: | 2003-03-07 | | Release date: | 2003-05-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Ligand-receptor binding revealed by the TNF family member TALL-1.
Nature, 423, 2003
|
|
1OQE
 
 | | Crystal structure of sTALL-1 with BAFF-R | | Descriptor: | Tumor necrosis factor ligand superfamily member 13B, soluble form, Tumor necrosis factor receptor superfamily member 13C | | Authors: | Zhang, G. | | Deposit date: | 2003-03-07 | | Release date: | 2003-05-13 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand-receptor binding revealed by the TNF family member TALL-1.
Nature, 423, 2003
|
|
5JKG
 
 | | The crystal structure of FGFR4 kinase domain in complex with LY2874455 | | Descriptor: | 2-[4-[E-2-[5-[(1R)-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-1H-indazol-3-yl]ethenyl]pyrazol-1-yl]ethanol, Fibroblast growth factor receptor 4 | | Authors: | Wu, D, Chen, L, Chen, Y. | | Deposit date: | 2016-04-26 | | Release date: | 2016-10-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | Crystal Structure of the FGFR4/LY2874455 Complex Reveals Insights into the Pan-FGFR Selectivity of LY2874455
Plos One, 11, 2016
|
|
4XFO
 
 | |
4XFN
 
 | |
5H7V
 
 | |
6PWS
 
 | |
6PWT
 
 | |
6PWR
 
 | |
7V63
 
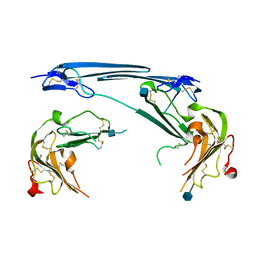 | | Structure of dimeric uPAR at low pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Urokinase plasminogen activator surface receptor | | Authors: | Yuan, C, Huang, M. | | Deposit date: | 2021-08-19 | | Release date: | 2021-12-22 | | Last modified: | 2022-04-06 | | Method: | X-RAY DIFFRACTION (2.906 Å) | | Cite: | Crystal structure and cellular functions of uPAR dimer
Nat Commun, 13, 2022
|
|
4ZNN
 
 | | MicroED structure of the segment, GVVHGVTTVA, from the A53T familial mutant of Parkinson's disease protein, alpha-synuclein residues 47-56 | | Descriptor: | Alpha-synuclein | | Authors: | Rodriguez, J.A, Ivanova, M, Sawaya, M.R, Cascio, D, Reyes, F, Shi, D, Johnson, L, Guenther, E, Sangwan, S, Hattne, J, Nannenga, B, Brewster, A.S, Messerschmidt, M, Boutet, S, Sauter, N.K, Gonen, T, Eisenberg, D.S. | | Deposit date: | 2015-05-05 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.41 Å) | | Cite: | Structure of the toxic core of alpha-synuclein from invisible crystals.
Nature, 525, 2015
|
|
7VNI
 
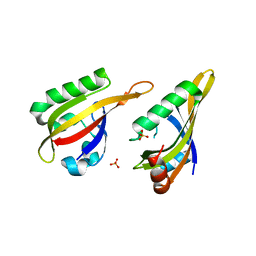 | | AHR-ARNT PAS-B heterodimer | | Descriptor: | Ahr homolog spineless, Aryl hydrocarbon receptor nuclear translocator, SULFATE ION | | Authors: | Dai, S.Y. | | Deposit date: | 2021-10-11 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural insight into the ligand binding mechanism of aryl hydrocarbon receptor.
Nat Commun, 13, 2022
|
|
7VNA
 
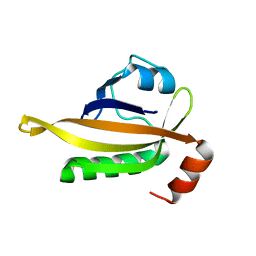 | | drosophlia AHR PAS-B domain | | Descriptor: | Ahr homolog spineless | | Authors: | Dai, S.Y. | | Deposit date: | 2021-10-10 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Structural insight into the ligand binding mechanism of aryl hydrocarbon receptor.
Nat Commun, 13, 2022
|
|
