3TGM
 
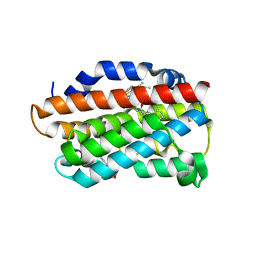 | | X-Ray Crystal Structure of Human Heme Oxygenase-1 in Complex with 1-(1H-imidazol-1-yl)-4,4-diphenyl-2 butanone | | Descriptor: | 1-(1H-imidazol-1-yl)-4,4-diphenylbutan-2-one, HEXANE-1,6-DIOL, Heme oxygenase 1, ... | | Authors: | Rahman, M.N, Jia, Z. | | Deposit date: | 2011-08-17 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A novel, "double-clamp" binding mode for human heme oxygenase-1 inhibition.
Plos One, 7, 2012
|
|
4ZME
 
 | |
4ZLH
 
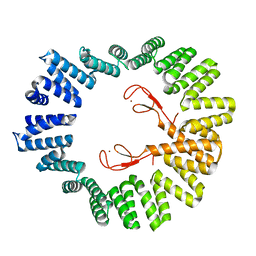 | | Structure of the LapB cytoplasmic domain at 2 angstroms | | Descriptor: | Lipopolysaccharide assembly protein B, ZINC ION | | Authors: | Prince, C.C, Jia, Z. | | Deposit date: | 2015-05-01 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An Unexpected Duo: Rubredoxin Binds Nine TPR Motifs to Form LapB, an Essential Regulator of Lipopolysaccharide Synthesis.
Structure, 23, 2015
|
|
4ZMF
 
 | |
4ZS4
 
 | |
4MF9
 
 | |
4MGF
 
 | |
4MLM
 
 | | Crystal Structure of PhnZ from uncultured bacterium HF130_AEPn_1 | | Descriptor: | FE (III) ION, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | van Staalduinen, L.M, McSorley, F.R, Zechel, D.L, Jia, Z, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2013-09-06 | | Release date: | 2014-04-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of PhnZ in complex with substrate reveals a di-iron oxygenase mechanism for catabolism of organophosphonates.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MLN
 
 | | Crystal of PhnZ bound to (R)-2-amino-1-hydroxyethylphosphonic acid | | Descriptor: | FE (III) ION, Predicted HD phosphohydrolase PhnZ, [(1R)-2-amino-1-hydroxyethyl]phosphonic acid | | Authors: | van Staalduinen, L.M, McSorley, F.R, Zechel, D.L, Jia, Z, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2013-09-06 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of PhnZ in complex with substrate reveals a di-iron oxygenase mechanism for catabolism of organophosphonates.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3CIO
 
 | |
4NUB
 
 | |
3DXM
 
 | | Structure of Bos taurus Arp2/3 Complex with Bound Inhibitor CK0993548 | | Descriptor: | (2S)-2-(3-bromophenyl)-3-(5-chloro-2-hydroxyphenyl)-1,3-thiazolidin-4-one, Actin-related protein 2, Actin-related protein 2/3 complex subunit 1B, ... | | Authors: | Nolen, B.J, Tomasevic, N, Russell, A, Pierce, D.W, Jia, Z, Hartman, J, Sakowicz, R, Pollard, T.D. | | Deposit date: | 2008-07-24 | | Release date: | 2009-07-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Characterization of two classes of small molecule inhibitors of Arp2/3 complex
Nature, 460, 2009
|
|
3DXK
 
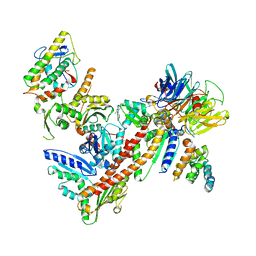 | | Structure of Bos Taurus Arp2/3 Complex with Bound Inhibitor CK0944636 | | Descriptor: | Actin-related protein 2, Actin-related protein 2/3 complex subunit 1B, Actin-related protein 2/3 complex subunit 2, ... | | Authors: | Nolen, B.J, Tomasevic, N, Russell, A, Pierce, D.W, Jia, Z, Hartman, J, Sakowicz, R, Pollard, T.D. | | Deposit date: | 2008-07-24 | | Release date: | 2009-07-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Characterization of two classes of small molecule inhibitors of Arp2/3 complex
Nature, 460, 2009
|
|
3DRW
 
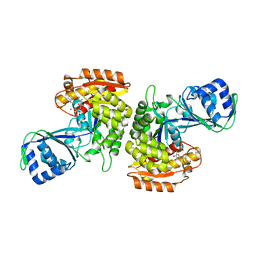 | | Crystal Structure of a Phosphofructokinase from Pyrococcus horikoshii OT3 with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADP-specific phosphofructokinase, SODIUM ION | | Authors: | Singer, A.U, Skarina, T, Kochinyan, S, Brown, G, Cuff, M.E, Edwards, A.M, Joachimiak, A, Savchenko, A, Yakunin, A.F, Jia, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-11 | | Release date: | 2008-12-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | ADP-dependent 6-phosphofructokinase from Pyrococcus horikoshii OT3: structure determination and biochemical characterization of PH1645.
J.Biol.Chem., 284, 2009
|
|
4P0U
 
 | |
3EPS
 
 | |
1NP8
 
 | | 18-k C-terminally trunucated small subunit of calpain | | Descriptor: | CADMIUM ION, Calcium-dependent protease, small subunit | | Authors: | Leinala, E.K, Arthur, J.S, Grochulski, P, Davies, P.L, Elce, J.S, Jia, Z. | | Deposit date: | 2003-01-17 | | Release date: | 2003-11-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A second binding site revealed by C-terminal truncation of calpain small subunit, a penta-EF-hand protein
PROTEINS: STRUCT.,FUNCT.,GENET., 53, 2003
|
|
1NT4
 
 | | Crystal structure of Escherichia coli periplasmic glucose-1-phosphatase H18A mutant complexed with glucose-1-phosphate | | Descriptor: | 1-O-phosphono-beta-D-glucopyranose, Glucose-1-phosphatase | | Authors: | Lee, D.C, Cottrill, M.A, Forsberg, C.W, Jia, Z, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2003-01-28 | | Release date: | 2004-01-13 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional insights revealed by the crystal structures of Escherichia coli glucose-1-phosphatase.
J.Biol.Chem., 278, 2003
|
|
1QXP
 
 | |
1R6Y
 
 | |
3G1P
 
 | |
6UTS
 
 | |
6UTU
 
 | | Crystal structure of minor pseudopilin ternary complex of XcpVWX from the Type 2 secretion system of Pseudomonas aeruginosa in the P3 space group | | Descriptor: | CALCIUM ION, Type II secretion system protein I, Type II secretion system protein J, ... | | Authors: | Zhang, Y, Wang, S, Jia, Z. | | Deposit date: | 2019-10-30 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | In Situ Proteolysis Condition-Induced Crystallization of the XcpVWX Complex in Different Lattices.
Int J Mol Sci, 21, 2020
|
|
3HOK
 
 | | X-ray Crystal Structure of Human Heme Oxygenase-1 with (2R, 4S)-2-[2-(4-Chlorophenyl)ethyl]-2-[(1H-imidazol-1-yl)methyl]-4[((5-trifluoromethylpyridin-2-yl)thio)methyl]-1,3-dioxolane: A Novel, Inducible Binding Mode | | Descriptor: | 2-({[(2R,4S)-2-[2-(4-chlorophenyl)ethyl]-2-(1H-imidazol-1-ylmethyl)-1,3-dioxolan-4-yl]methyl}sulfanyl)-5-(trifluoromethyl)pyridine, Heme oxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Rahman, M.N, Jia, Z. | | Deposit date: | 2009-06-02 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | X-ray crystal structure of human heme oxygenase-1 with (2R,4S)-2-[2-(4-chlorophenyl)ethyl]-2-[(1H-imidazol-1-yl)methyl]-4[((5-trifluoromethylpyridin-2-yl)thio)methyl]-1,3-dioxolane: a novel, inducible binding mode.
J.Med.Chem., 52, 2009
|
|
3I3L
 
 | | Crystal structure of CmlS, a flavin-dependent halogenase | | Descriptor: | Alkylhalidase CmlS, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Podzelinska, K, Soares, A, Jia, Z, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2009-06-30 | | Release date: | 2010-03-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chloramphenicol Biosynthesis: The Structure of CmlS, a Flavin-Dependent Halogenase Showing a Covalent Flavin-Aspartate Bond
J.Mol.Biol., 397, 2010
|
|
