6G7G
 
 | | Structure of SPH (Self-Incompatibility Protein Homologue) proteins, a widespread family of small, highly stable, secreted proteins from plants | | Descriptor: | S-protein homolog 15 | | Authors: | Rajasekar, K.V, Coulthard, R.J, Ride, J.P, Ji, S, Winn, P.J, Wheeler, M.P, Hyde, E.I, Smith, L.J. | | Deposit date: | 2018-04-06 | | Release date: | 2019-03-06 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structure of SPH (self-incompatibility protein homologue) proteins: a widespread family of small, highly stable, secreted proteins.
Biochem.J., 476, 2019
|
|
8ZB0
 
 | | Cryo-EM structure of human ZnT1 | | Descriptor: | Proton-coupled zinc antiporter SLC30A1, ZINC ION | | Authors: | Sun, S, Xie, E, Xu, S, Ji, S. | | Deposit date: | 2024-04-25 | | Release date: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The Intestinal Transporter SLC30A1 Plays a Critical Role in Regulating Systemic Zinc Homeostasis.
Adv Sci, 11, 2024
|
|
7XW9
 
 | | Cryo-EM structure of the TRH-bound human TRHR-Gq complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | Ji, S, Dong, Y, Chen, L, Zang, S, Shen, D, Guo, J, Qin, J, Zhang, H, Wang, W, Shen, Q, Mao, C, Zhang, Y. | | Deposit date: | 2022-05-26 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular basis for the activation of thyrotropin-releasing hormone receptor.
Cell Discov, 8, 2022
|
|
6E4R
 
 | | Crystal Structure of the Drosophila Melanogaster Polypeptide N-Acetylgalactosaminyl Transferase PGANT9B | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Samara, N.L, Tabak, L.A, Ten Hagen, K.G. | | Deposit date: | 2018-07-18 | | Release date: | 2018-09-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.061 Å) | | Cite: | A molecular switch orchestrates enzyme specificity and secretory granule morphology.
Nat Commun, 9, 2018
|
|
6E4Q
 
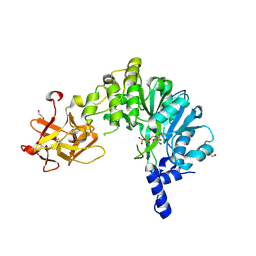 | | Crystal Structure of the Drosophila Melanogaster Polypeptide N-Acetylgalactosaminyl Transferase PGANT9A in Complex with UDP and Mn2+ | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Samara, N.L, Tabak, L.A, Ten Hagen, K.G. | | Deposit date: | 2018-07-18 | | Release date: | 2018-09-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | A molecular switch orchestrates enzyme specificity and secretory granule morphology.
Nat Commun, 9, 2018
|
|
7VBO
 
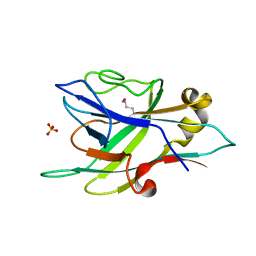 | | Alginate binding domain CBM | | Descriptor: | Alginate lyase, CALCIUM ION, SULFATE ION | | Authors: | Ji, S.Q, She, Q. | | Deposit date: | 2021-09-01 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification and structural analysis of a carbohydrate-binding module specific to alginate, a representative of a new family, CBM96.
J.Biol.Chem., 299, 2023
|
|
6JPH
 
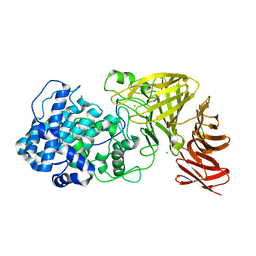 | | Crystal structure of the catalytic domain of a multi-domain alginate lyase Dp0100 from thermophilic bacterium Defluviitalea phaphyphila | | Descriptor: | ACETATE ION, Alginate lyase, CALCIUM ION, ... | | Authors: | Ji, S.Q, Dix, S.R, Aziz, A, Sedelnikova, S.E, Li, F.L, Rice, D.W. | | Deposit date: | 2019-03-27 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.759 Å) | | Cite: | The molecular basis of endolytic activity of a multidomain alginate lyase fromDefluviitalea phaphyphila, a representative of a new lyase family, PL39.
J.Biol.Chem., 294, 2019
|
|
6JPN
 
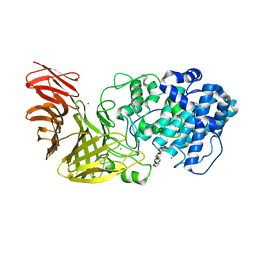 | | Crystal structure of the catalytic domain of a multi-domain alginate lyase Dp0100 from thermophilic bacterium Defluviitalea phaphyphila | | Descriptor: | Alginate lyase, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Ji, S.Q, Dix, S.R, Aziz, A, Sedelnikova, S.E, Li, F.L, Rice, D.W. | | Deposit date: | 2019-03-27 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The molecular basis of endolytic activity of a multidomain alginate lyase fromDefluviitalea phaphyphila, a representative of a new lyase family, PL39.
J.Biol.Chem., 294, 2019
|
|
6JP4
 
 | | Crystal structure of the catalytic domain of a multi-domain alginate lyase Dp0100 from thermophilic bacterium Defluviitalea phaphyphila | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Alginate lyase, ... | | Authors: | Ji, S.Q, Dix, S.R, Aziz, A, Sedelnikova, S.E, Li, F.L, Rice, D.W. | | Deposit date: | 2019-03-25 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.069 Å) | | Cite: | The molecular basis of endolytic activity of a multidomain alginate lyase fromDefluviitalea phaphyphila, a representative of a new lyase family, PL39.
J.Biol.Chem., 294, 2019
|
|
7XA9
 
 | | Structure of Arabidopsis thaliana CLCa | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chloride channel protein CLC-a, MAGNESIUM ION, ... | | Authors: | Ji, S, Jin, H, Kaiming, Z, Mingxing, W, Shanshan, L, Long, C. | | Deposit date: | 2022-03-17 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM structure of the plant nitrate transporter AtCLCa reveals characteristics of the anion-binding site and the ATP-binding pocket.
J.Biol.Chem., 299, 2023
|
|
8HU7
 
 | |
8HUE
 
 | | Crystal structure of FGF2-M2 mutant - D28E/C78I/C96I/S137P | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, Fibroblast growth factor 2 | | Authors: | Jung, Y.E, Cha, S.S, An, Y.J. | | Deposit date: | 2022-12-23 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural and biochemical investigation into stable FGF2 mutants with novel mutation sites and hydrophobic replacements for surface-exposed cysteines.
Plos One, 19, 2024
|
|
8IKJ
 
 | | Cryo-EM structure of the inactive CD97 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Adhesion G protein-coupled receptor E5,Soluble cytochrome b562,Adhesion G protein-coupled receptor E5 subunit beta | | Authors: | Mao, C, Zhao, R, Dong, Y, Gao, M, Chen, L, Zhang, C, Xiao, P, Guo, J, Qin, J, Shen, D, Ji, S, Zang, S, Zhang, H, Wang, W, Shen, Q, Sun, P, Zhang, Y. | | Deposit date: | 2023-02-28 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Conformational transitions and activation of the adhesion receptor CD97.
Mol.Cell, 84, 2024
|
|
7CIS
 
 | | Peptide modification of MHC class I molecules | | Descriptor: | ARG-ARG-PHE-SEP-ARG-SEP-PRO-ILE-ARG, Beta-2-microglobulin, MHC class I antigen | | Authors: | Sun, M.W, Feng, L, Qi, J.X, Liu, W.J, Gao, G.F. | | Deposit date: | 2020-07-08 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phosphosite-dependent presentation of dual phosphorylated peptides by MHC class I molecules.
Iscience, 25, 2022
|
|
7CIR
 
 | | Peptide phosphorylation modification of MHC class I molecules | | Descriptor: | ARG-ARG-PHE-SEP-ARG-SER-PRO-ILE-ARG-ARG, Beta-2-microglobulin, MHC class I antigen | | Authors: | Sun, M.W, Feng, L, Qi, J.X, Liu, W.J, Gao, G.F. | | Deposit date: | 2020-07-08 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Phosphosite-dependent presentation of dual phosphorylated peptides by MHC class I molecules.
Iscience, 25, 2022
|
|
7CIQ
 
 | | Phosphorylation modification of MHC I polypeptide | | Descriptor: | ARG-ARG-PHE-SER-ARG-SER-PRO-ILE-ARG-ARG, Beta-2-microglobulin, MHC class I antigen | | Authors: | Sun, M.W, Feng, L, Qi, J.X, Liu, W.J, Gao, G.F. | | Deposit date: | 2020-07-08 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Phosphosite-dependent presentation of dual phosphorylated peptides by MHC class I molecules.
Iscience, 25, 2022
|
|
8XZG
 
 | | Cryo-EM structure of the [Pyr1]-apelin-13-bound human APLNR-Gi complex | | Descriptor: | Apelin receptor, Apelin-13, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wang, W, Ji, S, Zhang, Y. | | Deposit date: | 2024-01-21 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
8XZF
 
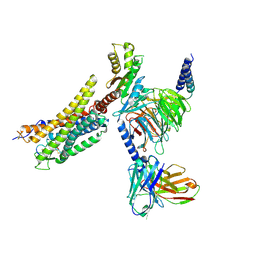 | | Cryo-EM structure of the WN561-bound human APLNR-Gi complex | | Descriptor: | Apelin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, W, Ji, S, Zhang, Y. | | Deposit date: | 2024-01-21 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
8XZJ
 
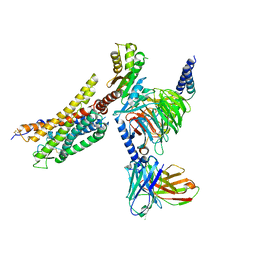 | | Cryo-EM structure of the WN353-bound human APLNR-Gi complex | | Descriptor: | Apelin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, W, Ji, S, Zhang, Y. | | Deposit date: | 2024-01-21 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
8XZH
 
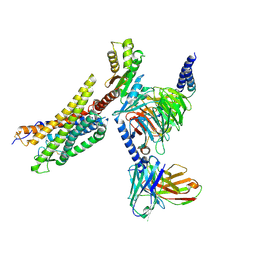 | | Cryo-EM structure of the MM07-bound human APLNR-Gi complex | | Descriptor: | Apelin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, W, Ji, S, Zhang, Y. | | Deposit date: | 2024-01-21 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
8XZI
 
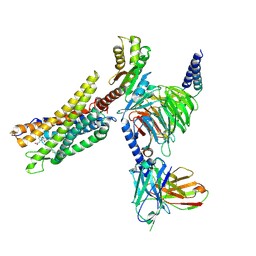 | | Cryo-EM structure of the CMF-019-bound human APLNR-Gi complex | | Descriptor: | (3~{S})-5-methyl-3-[[1-pentan-3-yl-2-(thiophen-2-ylmethyl)benzimidazol-5-yl]carbonylamino]hexanoic acid, Apelin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wang, W, Ji, S, Zhang, Y. | | Deposit date: | 2024-01-21 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
8Z40
 
 | |
8Z3K
 
 | | The structure of type III CRISPR-associated deaminase in complex 2cA6-2ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenosine deaminase domain-containing protein, RNA (5'-R(P*AP*AP*AP*AP*AP*A)-3'), ... | | Authors: | Chen, M.R, Li, Z.X, Xiao, Y.B. | | Deposit date: | 2024-04-15 | | Release date: | 2024-12-11 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Antiviral signaling of a type III CRISPR-associated deaminase.
Science, 387, 2025
|
|
8Z3R
 
 | | The structure of type III CRISPR-associated deaminase in complex cA4 | | Descriptor: | 3'-O-[(R)-{[(2S,3aS,4S,6S,6aS)-6-(6-amino-9H-purin-9-yl)-2-hydroxy-2-oxotetrahydro-2H-2lambda~5~-furo[3,4-d][1,3,2]dioxaphosphol-4-yl]methoxy}(hydroxy)phosphoryl]adenosine, Adenosine deaminase domain-containing protein, ZINC ION | | Authors: | Chen, M.R, Li, Z.X, Xiao, Y.B. | | Deposit date: | 2024-04-16 | | Release date: | 2024-12-25 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (2.28 Å) | | Cite: | Antiviral signaling of a type III CRISPR-associated deaminase.
Science, 387, 2025
|
|
8Z3P
 
 | | The structure of type III CRISPR-associated deaminase in complex cA6 and ATP, fully activated | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenosine deaminase domain-containing protein, MAGNESIUM ION, ... | | Authors: | Chen, M.R, Li, Z.X, Xiao, Y.B. | | Deposit date: | 2024-04-15 | | Release date: | 2024-12-25 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Antiviral signaling of a type III CRISPR-associated deaminase.
Science, 387, 2025
|
|
