3O2A
 
 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | GLUTAMIC ACID, GLYCEROL, Glutamate receptor 2, ... | | Authors: | Maclean, J.K.F, Basten, S, Campbell, R.A, Cumming, I.A, Gillen, K.J, Gillespie, J, Jamieson, C, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Moir, E.M, Morrow, J.A, Papakosta, M, Rankovic, Z, Smith, L. | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-15 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel series of positive modulators of the AMPA receptor: discovery and structure based hit-to-lead studies.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3O6I
 
 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | 2-[({3-tert-butyl-4-[(methylamino)methyl]-1H-pyrazol-1-yl}acetyl)amino]-4,5,6,7-tetrahydro-1-benzothiophene-3-carboxamide, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Maclean, J.K.F, Campbell, R.A, Cumming, I.A, Gillen, K.J, Gillespie, J, Jamieson, C, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Martin, F, Moir, E.M, Morrow, J.A, Pantling, J, Rankovic, Z, Smith, L. | | Deposit date: | 2010-07-29 | | Release date: | 2010-09-15 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel series of positive modulators of the AMPA receptor: structure-based lead optimization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3O29
 
 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | GLUTAMIC ACID, GLYCEROL, Glutamate receptor 2, ... | | Authors: | Maclean, J.K.F, Basten, S, Campbell, R.A, Cumming, I.A, Gillen, K.J, Gillespie, J, Jamieson, C, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Moir, E.M, Morrow, J.A, Papakosta, M, Rankovic, Z, Smith, L. | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-15 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A novel series of positive modulators of the AMPA receptor: discovery and structure based hit-to-lead studies.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3O28
 
 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | 2-({[3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl]acetyl}amino)-4,5,6,7-tetrahydro-1-benzothiophene-3-carboxamide, DIMETHYL SULFOXIDE, GLUTAMIC ACID, ... | | Authors: | Maclean, J.K.F, Basten, S, Campbell, R.A, Cumming, I.A, Gillen, K.J, Gillespie, J, Jamieson, C, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Moir, E.M, Morrow, J.A, Papakosta, M, Rankovic, Z, Smith, L. | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-15 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel series of positive modulators of the AMPA receptor: discovery and structure based hit-to-lead studies.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3O6G
 
 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | 1,2-ETHANEDIOL, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Maclean, J.K.F, Campbell, R.A, Cumming, I.A, Gillen, K.J, Gillespie, J, Jamieson, C, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Martin, F, Moir, E.M, Morrow, J.A, Pantling, J, Rankovic, Z, Smith, L. | | Deposit date: | 2010-07-29 | | Release date: | 2010-09-15 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel series of positive modulators of the AMPA receptor: structure-based lead optimization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3O6H
 
 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | 1,2-ETHANEDIOL, 2-[({4-[(ethylamino)methyl]-3-(trifluoromethyl)-1H-pyrazol-1-yl}acetyl)amino]-4,5,6,7-tetrahydro-1-benzothiophene-3-carboxamide, DIMETHYL SULFOXIDE, ... | | Authors: | Maclean, J.K.F, Campbell, R.A, Cumming, I.A, Gillen, K.J, Gillespie, J, Jamieson, C, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Martin, F, Moir, E.M, Morrow, J.A, Pantling, J, Rankovic, Z, Smith, L. | | Deposit date: | 2010-07-29 | | Release date: | 2010-09-15 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A novel series of positive modulators of the AMPA receptor: structure-based lead optimization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3PMW
 
 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | DIMETHYL SULFOXIDE, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Maclean, J.K.F, Jamieson, C, Brown, C.I, Campbell, R.A, Gillen, K.J, Gillespie, J, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Moir, E.M, Morrow, J.A, Pantling, J, Rankovic, Z, Smith, L. | | Deposit date: | 2010-11-18 | | Release date: | 2011-01-12 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure based evolution of a novel series of positive modulators of the AMPA receptor.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3PMX
 
 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, GLUTAMIC ACID, ... | | Authors: | Maclean, J.K.F, Jamieson, C, Brown, C.I, Campbell, R.A, Gillen, K.J, Gillespie, J, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Moir, E.M, Morrow, J.A, Pantling, J, Rankovic, Z, Smith, L. | | Deposit date: | 2010-11-18 | | Release date: | 2011-01-12 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure based evolution of a novel series of positive modulators of the AMPA receptor.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3PMV
 
 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | GLUTAMIC ACID, GLYCEROL, Glutamate receptor 2, ... | | Authors: | Maclean, J.K.F, Jamieson, C, Brown, C.I, Campbell, R.A, Gillen, K.J, Gillespie, J, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Moir, E.M, Morrow, J.A, Pantling, J, Rankovic, Z, Smith, L. | | Deposit date: | 2010-11-18 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure based evolution of a novel series of positive modulators of the AMPA receptor.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
7Z0N
 
 | | Structure-Based Design of a Novel Class of Autotaxin Inhibitors Based on Endogenous Allosteric Modulators | | Descriptor: | CALCIUM ION, GLYCEROL, IODIDE ION, ... | | Authors: | Salgado-Polo, F, Clark, J.M, Macdonald, S.J.F, Barrett, T.N, Perrakis, A, Jamieson, A. | | Deposit date: | 2022-02-23 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Design of a Novel Class of Autotaxin Inhibitors Based on Endogenous Allosteric Modulators.
J.Med.Chem., 65, 2022
|
|
5I5S
 
 | |
5I5V
 
 | | X-RAY CRYSTAL STRUCTURE AT 1.94A RESOLUTION OF HUMAN MITOCHONDRIAL BRANCHED CHAIN AMINOTRANSFERASE (BCATM) COMPLEXED WITH A THIENOPYRIMIDINE COMPOUND AND AN INTERNAL ALDIMINE LINKED PLP COFACTOR. | | Descriptor: | 1,2-ETHANEDIOL, 3,5-dimethyl-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidine-6-carboxylic acid, Branched-chain-amino-acid aminotransferase, ... | | Authors: | Somers, D.O. | | Deposit date: | 2016-02-15 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structurally Diverse Mitochondrial Branched Chain Aminotransferase (BCATm) Leads with Varying Binding Modes Identified by Fragment Screening.
J.Med.Chem., 59, 2016
|
|
5I5T
 
 | | X-RAY CRYSTAL STRUCTURE AT 2.31A RESOLUTION OF HUMAN MITOCHONDRIAL BRANCHED CHAIN AMINOTRANSFERASE (BCATM) COMPLEXED WITH A TETRAHYDROQUINOLINE COMPOUND AND AN INTERNAL ALDIMINE LINKED PLP COFACTOR. | | Descriptor: | (3R)-3-methyl-1,2,3,4-tetrahydroquinoline-8-sulfonamide, 1,2-ETHANEDIOL, Branched-chain-amino-acid aminotransferase, ... | | Authors: | Somers, D.O. | | Deposit date: | 2016-02-15 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structurally Diverse Mitochondrial Branched Chain Aminotransferase (BCATm) Leads with Varying Binding Modes Identified by Fragment Screening.
J.Med.Chem., 59, 2016
|
|
5I5Y
 
 | |
5I60
 
 | |
3KWB
 
 | | Structure of CatK covalently bound to a dioxo-triazine inhibitor | | Descriptor: | 3,5-dioxo-4-(3-piperidin-1-ylpropyl)-2-[3-(trifluoromethyl)phenyl]-2,3,4,5-tetrahydro-1,2,4-triazine-6-carbonitrile, Cathepsin K | | Authors: | Uitdehaag, J.C.M, van Zeeland, M. | | Deposit date: | 2009-12-01 | | Release date: | 2010-04-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Dioxo-triazines as a novel series of cathepsin K inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5I5X
 
 | | X-RAY CRYSTAL STRUCTURE AT 1.65A RESOLUTION OF HUMAN MITOCHONDRIAL BRANCHED CHAIN AMINOTRANSFERASE (BCATM) COMPLEXED WITH A THIAZOLE COMPOUND AND PMP COFACTOR. | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 5-methyl-4-oxo-N-(1,3,4-thiadiazol-2-yl)-3,4-dihydrothieno[2,3-d]pyrimidine-6-carboxamide, ... | | Authors: | Somers, D.O. | | Deposit date: | 2016-02-15 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structurally Diverse Mitochondrial Branched Chain Aminotransferase (BCATm) Leads with Varying Binding Modes Identified by Fragment Screening.
J.Med.Chem., 59, 2016
|
|
5I5W
 
 | |
5I5U
 
 | | X-RAY CRYSTAL STRUCTURE AT 2.40A RESOLUTION OF HUMAN MITOCHONDRIAL BRANCHED CHAIN AMINOTRANSFERASE (BCATM) COMPLEXED WITH A TETRAHYDRONAPHTHALENYL COMPOUND AND AN INTERNAL ALDIMINE LINKED PLP COFACTOR. | | Descriptor: | 1,2-ETHANEDIOL, 2-hydroxy-N-[(1R)-1,2,3,4-tetrahydronaphthalen-1-yl]acetamide, Branched-chain-amino-acid aminotransferase, ... | | Authors: | Somers, D.O. | | Deposit date: | 2016-02-15 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | Structurally Diverse Mitochondrial Branched Chain Aminotransferase (BCATm) Leads with Varying Binding Modes Identified by Fragment Screening.
J.Med.Chem., 59, 2016
|
|
8B5G
 
 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH 7,8-dimethoxy-3-methyl-1,3-dihydro-2H-benzo[d]azepin-2-one | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 7,8-dimethoxy-3-methyl-1~{H}-3-benzazepin-2-one, ... | | Authors: | Chung, C. | | Deposit date: | 2022-09-22 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.619 Å) | | Cite: | Identification and Optimization of a Ligand-Efficient Benzoazepinone Bromodomain and Extra Terminal (BET) Family Acetyl-Lysine Mimetic into the Oral Candidate Quality Molecule I-BET432.
J.Med.Chem., 65, 2022
|
|
8B5H
 
 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH (R)-7-((R)-1,2-dihydroxyethyl)-1,3-dimethyl-5-(1-methyl-1H-pyrazol-4-yl)-1,3-dihydro-2H-benzo[d]azepin-2-one | | Descriptor: | (1~{R})-7-[(1~{R})-1,2-bis(oxidanyl)ethyl]-1,3-dimethyl-5-(1-methylpyrazol-4-yl)-1~{H}-3-benzazepin-2-one, 1,2-ETHANEDIOL, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, ... | | Authors: | Chung, C. | | Deposit date: | 2022-09-22 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Identification and Optimization of a Ligand-Efficient Benzoazepinone Bromodomain and Extra Terminal (BET) Family Acetyl-Lysine Mimetic into the Oral Candidate Quality Molecule I-BET432.
J.Med.Chem., 65, 2022
|
|
8B5I
 
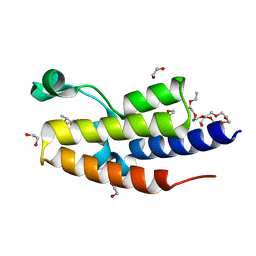 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH 7,8-dimethoxy-1,3-dimethyl-1,3-dihydro-2H-benzo[d]azepin-2-one | | Descriptor: | (1~{R})-7,8-dimethoxy-1,3-dimethyl-1~{H}-3-benzazepin-2-one, 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Chung, C. | | Deposit date: | 2022-09-22 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Identification and Optimization of a Ligand-Efficient Benzoazepinone Bromodomain and Extra Terminal (BET) Family Acetyl-Lysine Mimetic into the Oral Candidate Quality Molecule I-BET432.
J.Med.Chem., 65, 2022
|
|
8B5J
 
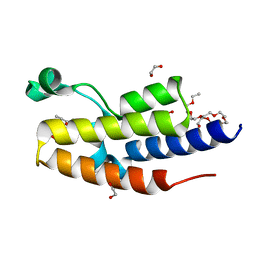 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH 7,8-dimethoxy-1,3-dimethyl-1,3-dihydro-2H-benzo[d]azepin-2-one | | Descriptor: | (5~{R})-7,8-dimethoxy-3,5-dimethyl-2,5-dihydro-1~{H}-3-benzazepin-4-one, 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Chung, C. | | Deposit date: | 2022-09-22 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Identification and Optimization of a Ligand-Efficient Benzoazepinone Bromodomain and Extra Terminal (BET) Family Acetyl-Lysine Mimetic into the Oral Candidate Quality Molecule I-BET432.
J.Med.Chem., 65, 2022
|
|
1OGW
 
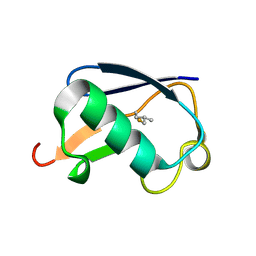 | | Synthetic Ubiquitin with fluoro-Leu at 50 and 67 | | Descriptor: | UBIQUITIN | | Authors: | Alexeev, D, Ramage, R, Young, D.W, Sawyer, L. | | Deposit date: | 2003-05-13 | | Release date: | 2003-05-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Synthesis, Structural and Biological Studies of Ubiquitin Mutants Containing (2S, 4S)-5-Fluoroleucine Residues Strategically Placed in the Hydrophobic Core
Chembiochem, 4, 2003
|
|
6ZAD
 
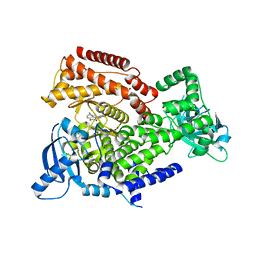 | | PI3K Delta in complex with methoxymethyloxathiatetraazatetracyclodocosahexaenedione | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, methoxymethyloxathiatetraazatetracyclodocosahexaenedione | | Authors: | Convery, M.A, Hardy, C.J, Spencer, J.A, Rowland, P. | | Deposit date: | 2020-06-05 | | Release date: | 2020-07-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Design and Development of a Macrocyclic Series Targeting Phosphoinositide 3-Kinase delta.
Acs Med.Chem.Lett., 11, 2020
|
|
