6DDA
 
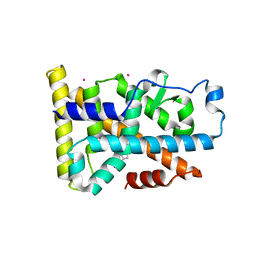 | | Nurr1 Covalently Modified by a Dopamine Metabolite | | Descriptor: | 5-hydroxy-1,2-dihydro-6H-indol-6-one, BROMIDE ION, Nuclear receptor subfamily 4 group A member 2, ... | | Authors: | Bruning, J.M, Wang, Y, Otrabella, F, Boxue, T, Liu, H, Bhattacharya, P, Guo, S, Holton, J.M, Fletterick, R.J, Jacobson, M.P, England, P.M. | | Deposit date: | 2018-05-09 | | Release date: | 2019-03-20 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Covalent Modification and Regulation of the Nuclear Receptor Nurr1 by a Dopamine Metabolite.
Cell Chem Biol, 26, 2019
|
|
3LVG
 
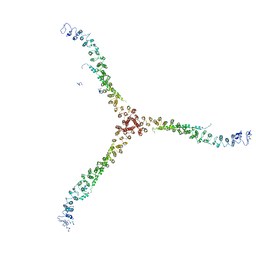 | | Crystal structure of a clathrin heavy chain and clathrin light chain complex | | Descriptor: | Clathrin heavy chain 1, Clathrin light chain B | | Authors: | Wilbur, J.D, Hwang, P.K, Ybe, J.A, Lane, M, Sellers, B.D, Jacobson, M.P, Fletterick, R.J, Brodsky, F.M. | | Deposit date: | 2010-02-20 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (7.94 Å) | | Cite: | Conformation switching of clathrin light chain regulates clathrin lattice assembly.
Dev.Cell, 18, 2010
|
|
3LVH
 
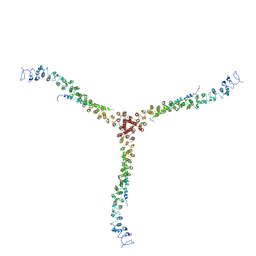 | | Crystal structure of a clathrin heavy chain and clathrin light chain complex | | Descriptor: | Clathrin heavy chain 1, Clathrin light chain B | | Authors: | Wilbur, J.D, Hwang, P.K, Ybe, J.A, Lane, M, Sellers, B.D, Jacobson, M.P, Fletterick, R.J, Brodsky, F.M. | | Deposit date: | 2010-02-20 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (9 Å) | | Cite: | Conformation switching of clathrin light chain regulates clathrin lattice assembly.
Dev.Cell, 18, 2010
|
|
4MEV
 
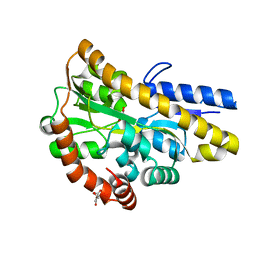 | | Crystal structure of a TRAP periplasmic solute binding protein from Rhodoferax ferrireducens (Rfer_1840), Target EFI-510211, with bound malonate, space group I422 | | Descriptor: | CITRIC ACID, MALONATE ION, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Zhao, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4MCO
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Rhodoferax ferrireducens (Rfer_1840), target EFI-510211, with bound malonate | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MALONATE ION, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-21 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4MNC
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Polaromonas sp. JS666 (Bpro_4736), Target EFI-510156, with bound benzoyl formate, space group P21 | | Descriptor: | BENZOYL-FORMIC ACID, SULFATE ION, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-10 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4NAV
 
 | | Crystal structure of hypothetical protein XCC2798 from Xanthomonas campestris, Target EFI-508608 | | Descriptor: | HYPOTHETICAL PROTEIN XCC279 | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Glenn, A.S, Chowdhury, S, Evans, B, Zhao, S.C, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Stead, M, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-22 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of hypothetical protein XCC2798 from Xanthomonas campestris, Target EFI-508608
TO BE PUBLISHED
|
|
4MIJ
 
 | | Crystal structure of a Trap periplasmic solute binding protein from Polaromonas sp. JS666 (Bpro_3107), target EFI-510173, with bound alpha/beta D-Galacturonate, space group P21 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-31 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4MNI
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Polaromonas sp. JS666 (Bpro_4736), Target EFI-510156, with bound benzoyl formate, space group P6522 | | Descriptor: | BENZOYL-FORMIC ACID, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-10 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4MHF
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Polaromonas sp. JS666 (Bpro_3107), target EFI-510173, with bound alpha/beta D-Glucuronate, space group P21 | | Descriptor: | TRAP dicarboxylate transporter, DctP subunit, alpha-D-glucopyranuronic acid, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-29 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4N0V
 
 | | Crystal structure of a glutathione S-transferase domain-containing protein (Marinobacter aquaeolei VT8), Target EFI-507332 | | Descriptor: | Glutathione S-transferase, N-terminal domain | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Glenn, A.S, Chowdhury, S, Evans, B, Zhao, S.C, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Stead, M, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-02 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a glutathione S-transferase domain-containing protein (Marinobacter aquaeolei VT8), Target EFI-507332
TO BE PUBLISHED
|
|
4N4U
 
 | | Crystal structure of ABC transporter solute binding protein BB0719 from Bordetella bronchiseptica RB50, TARGET EFI-510049 | | Descriptor: | GLYCEROL, Putative ABC transporter periplasmic solute-binding protein | | Authors: | Patskovsky, Y, Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-08 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4N17
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Burkholderia ambifaria (BAM_6123), Target EFI-510059, With bound beta-D-galacturonate | | Descriptor: | CALCIUM ION, CHLORIDE ION, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Zhao, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-03 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4MX6
 
 | | Crystal structure of a trap periplasmic solute binding protein from shewanella oneidensis (SO_3134), target EFI-510275, with bound succinate | | Descriptor: | CHLORIDE ION, SUCCINIC ACID, TRAP-type C4-dicarboxylate:H+ symport system substrate-binding component DctP | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-25 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4N15
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Burkholderia ambifaria (BAM_6123), Target EFI-510059, with bound beta-D-glucuronate | | Descriptor: | MAGNESIUM ION, TRAP dicarboxylate transporter, DctP subunit, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Zhao, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-03 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4UAB
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Chromohalobacter salexigens DSM 3043 (Csal_0678), Target EFI-501078, with bound ethanolamine | | Descriptor: | CHLORIDE ION, ETHANOLAMINE, Twin-arginine translocation pathway signal | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-08-08 | | Release date: | 2014-09-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
3LOM
 
 | | CRYSTAL STRUCTURE OF GERANYLTRANSFERASE FROM Legionella pneumophila | | Descriptor: | Geranyltranstransferase, PHOSPHATE ION | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-04 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3UCA
 
 | | Crystal structure of isoprenoid synthase (target EFI-501974) from clostridium perfringens | | Descriptor: | Geranyltranstransferase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-10-26 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4FP4
 
 | | Crystal structure of isoprenoid synthase a3mx09 (target efi-501993) from pyrobaculum calidifontis | | Descriptor: | GERAN-8-YL GERAN, Polyprenyl synthetase, UNKNOWN ATOM OR ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-06-21 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4F62
 
 | | Crystal structure of a putative farnesyl-diphosphate synthase from Marinomonas sp. MED121 (Target EFI-501980) | | Descriptor: | CHLORIDE ION, GLYCEROL, Geranyltranstransferase, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-05-14 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3DER
 
 | | Crystal structure of dipeptide epimerase from Thermotoga maritima complexed with L-Ala-L-Lys dipeptide | | Descriptor: | ALANINE, LYSINE, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Imker, H.J, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2008-06-10 | | Release date: | 2008-11-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a dipeptide epimerase enzymatic function guided by homology modeling and virtual screening.
Structure, 16, 2008
|
|
3DEQ
 
 | | Crystal structure of dipeptide epimerase from Thermotoga maritima complexed with L-Ala-L-Leu dipeptide | | Descriptor: | ALANINE, LEUCINE, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Imker, H.J, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2008-06-10 | | Release date: | 2008-11-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a dipeptide epimerase enzymatic function guided by homology modeling and virtual screening.
Structure, 16, 2008
|
|
3DFY
 
 | | Crystal structure of apo dipeptide epimerase from Thermotoga maritima | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Fedorov, A.A, Fedorov, E.V, Imker, H.J, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2008-06-12 | | Release date: | 2008-11-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a dipeptide epimerase enzymatic function guided by homology modeling and virtual screening.
Structure, 16, 2008
|
|
7W6P
 
 | | Cryo-EM structure of the alpha2A adrenergic receptor GoA signaling complex bound to a G protein biased agonist | | Descriptor: | Alpha-2A adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, J, Fink, E.A, Shoichet, B.K, Du, Y. | | Deposit date: | 2021-12-02 | | Release date: | 2022-09-28 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structure-based discovery of nonopioid analgesics acting through the alpha 2A -adrenergic receptor.
Science, 377, 2022
|
|
7W7E
 
 | | Cryo-EM structure of the alpha2A adrenergic receptor GoA signaling complex bound to a biased agonist | | Descriptor: | 5-(3-bicyclo[4.2.0]octa-1,3,5-trienyl)-1,2,3,6-tetrahydropyridine, Alpha-2A adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, J, Fink, E.A, Shoichet, B.K, Du, Y. | | Deposit date: | 2021-12-04 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure-based discovery of nonopioid analgesics acting through the alpha 2A -adrenergic receptor.
Science, 377, 2022
|
|
