4PNY
 
 | |
4PMC
 
 | |
4PMB
 
 | |
7QDH
 
 | | SARS-CoV-2 S protein S:D614G mutant 1-up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Ginex, T, Marco-Marin, C, Wieczor, M, Mata, C.P, Krieger, J, Lopez-Redondo, M.L, Frances-Gomez, C, Ruiz-Rodriguez, P, Melero, R, Sanchez-Sorzano, C.O, Martinez, M, Gougeard, N, Forcada-Nadal, A, Zamora-Caballero, S, Gozalbo-Rovira, R, Sanz-Frasquet, C, Bravo, J, Rubio, V, Marina, A, Geller, R, Comas, I, Gil, C, Coscolla, M, Orozco, M, LLacer, J.L, Carazo, J.M. | | Deposit date: | 2021-11-27 | | Release date: | 2022-05-25 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The structural role of SARS-CoV-2 genetic background in the emergence and success of spike mutations: The case of the spike A222V mutation.
Plos Pathog., 18, 2022
|
|
7QDG
 
 | | SARS-CoV-2 S protein S:A222V + S:D614G mutant 1-up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ginex, T, Marco-Marin, C, Wieczor, M, Mata, C.P, Krieger, J, Lopez-Redondo, M.L, Frances-Gomez, C, Ruiz-Rodriguez, P, Melero, R, Sanchez-Sorzano, C.O, Martinez, M, Gougeard, N, Forcada-Nadal, A, Zamora-Caballero, S, Gozalbo-Rovira, R, Sanz-Frasquet, C, Bravo, J, Rubio, V, Marina, A, Geller, R, Comas, I, Gil, C, Coscolla, M, Orozco, M, LLacer, J.L, Carazo, J.M. | | Deposit date: | 2021-11-27 | | Release date: | 2022-05-25 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The structural role of SARS-CoV-2 genetic background in the emergence and success of spike mutations: The case of the spike A222V mutation.
Plos Pathog., 18, 2022
|
|
7QTV
 
 | | Beryllium fluoride form of the Na+,K+-ATPase (E2-BeFx) | | Descriptor: | 1-O-decanoyl-beta-D-tagatofuranosyl beta-D-allopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fruergaard, M.U, Dach, I, Andersen, J.L, Ozol, M, Shahsavar, A, Quistgaard, E.M, Poulsen, H, Fedosova, N.U, Nissen, P. | | Deposit date: | 2022-01-16 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4.05 Å) | | Cite: | The Na + ,K + -ATPase in complex with beryllium fluoride mimics an ATPase phosphorylated state.
J.Biol.Chem., 298, 2022
|
|
2ACE
 
 | | NATIVE ACETYLCHOLINESTERASE (E.C. 3.1.1.7) FROM TORPEDO CALIFORNICA | | Descriptor: | ACETYLCHOLINE, ACETYLCHOLINESTERASE | | Authors: | Harel, M, Raves, M.L, Silman, I, Sussman, J.L. | | Deposit date: | 1996-06-23 | | Release date: | 1996-11-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of acetylcholinesterase complexed with the nootropic alkaloid, (-)-huperzine A.
Nat.Struct.Biol., 4, 1997
|
|
4TRD
 
 | |
1A2J
 
 | | OXIDIZED DSBA CRYSTAL FORM II | | Descriptor: | DISULFIDE BOND FORMATION PROTEIN | | Authors: | Martin, J.L, Guddat, L.W. | | Deposit date: | 1998-01-06 | | Release date: | 1998-09-16 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of reduced and oxidized DsbA: investigation of domain motion and thiolate stabilization.
Structure, 6, 1998
|
|
1A2L
 
 | | REDUCED DSBA AT 2.7 ANGSTROMS RESOLUTION | | Descriptor: | DISULFIDE BOND FORMATION PROTEIN | | Authors: | Martin, J.L, Guddat, L.W. | | Deposit date: | 1998-01-06 | | Release date: | 1998-07-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of reduced and oxidized DsbA: investigation of domain motion and thiolate stabilization.
Structure, 6, 1998
|
|
6O6W
 
 | | Solution structure of human myeloid-derived growth factor | | Descriptor: | Myeloid-derived growth factor | | Authors: | Bortnov, V, Tonelli, M, Lee, W, Markley, J.L, Mosher, D.F. | | Deposit date: | 2019-03-07 | | Release date: | 2019-11-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human myeloid-derived growth factor suggests a conserved function in the endoplasmic reticulum.
Nat Commun, 10, 2019
|
|
1EXV
 
 | | HUMAN LIVER GLYCOGEN PHOSPHORYLASE A COMPLEXED WITH GLCNAC AND CP-403,700 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, LIVER GLYCOGEN PHOSPHORYLASE, N-acetyl-beta-D-glucopyranosylamine, ... | | Authors: | Rath, V.L, Ammirati, M, Danley, D.E, Ekstrom, J.L, Hynes, T.R, Olson, T.V, Hoover, D.J. | | Deposit date: | 2000-05-04 | | Release date: | 2000-11-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human liver glycogen phosphorylase inhibitors bind at a new allosteric site.
Chem.Biol., 7, 2000
|
|
2OXJ
 
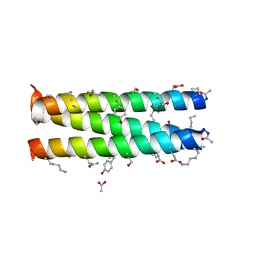 | | Helix Bundle Quaternary Structure from alpha/beta-Peptide Foldamers: GCN4-p1 with beta-residues at b and f heptad positions. | | Descriptor: | ACETATE ION, hybrid alpha/beta peptide based on the GCN4-p1 sequence; heptad positions b and f substituted with beta-amino acids | | Authors: | Horne, W.S, Price, J.L, Keck, J.L, Gellman, S.H. | | Deposit date: | 2007-02-20 | | Release date: | 2007-03-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Helix Bundle Quaternary Structure from alpha/beta-Peptide Foldamers.
J.Am.Chem.Soc., 129, 2007
|
|
2OXK
 
 | | Helix Bundle Quaternary Structure from alpha/beta-Peptide Foldamers: GCN4-pLI with beta-residues at b and f heptad positions. | | Descriptor: | FORMIC ACID, hybrid alpha/beta peptide based on the GCN4-pLI sequence; heptad positions b and f substituted with beta-amino acids | | Authors: | Horne, W.S, Price, J.L, Keck, J.L, Gellman, S.H. | | Deposit date: | 2007-02-20 | | Release date: | 2007-03-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Helix Bundle Quaternary Structure from alpha/beta-Peptide Foldamers.
J.Am.Chem.Soc., 129, 2007
|
|
4ZQ3
 
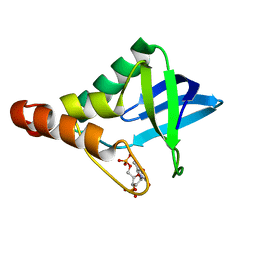 | |
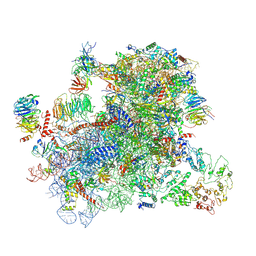
6GSN
 
 | |
4ZUJ
 
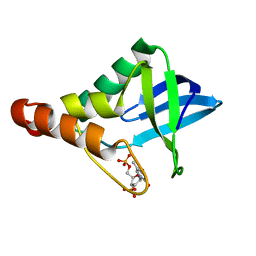 | |
6GSM
 
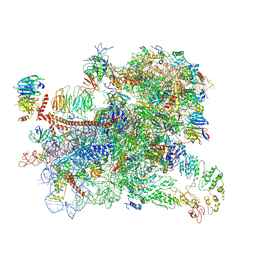 | | Structure of a partial yeast 48S preinitiation complex in open conformation. | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0, 40S ribosomal protein S1, ... | | Authors: | Llacer, J.L, Hussain, T, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2018-06-14 | | Release date: | 2019-07-31 | | Last modified: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Large-scale movement of eIF3 domains during translation initiation modulate start codon selection.
Nucleic Acids Res., 2021
|
|
4ZUI
 
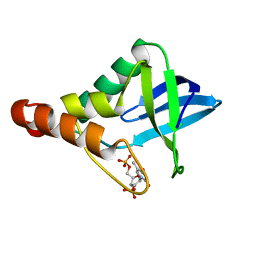 | |
1FVJ
 
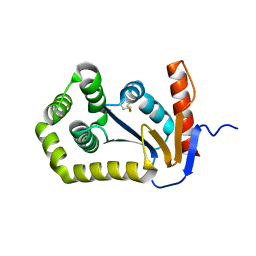 | |
6VA3
 
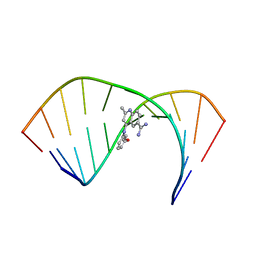 | | Solution Structure of the Tau pre-mRNA Exon 10 Splicing Regulatory Element Bound to MQC | | Descriptor: | 4-[(3-methoxyphenyl)amino]-2-methylquinoline-6-carboximidamide, RNA (5'-R(*CP*AP*CP*AP*CP*GP*UP*CP*GP*G)-3'), RNA (5'-R(*CP*CP*GP*GP*CP*AP*GP*UP*GP*UP*G)-3') | | Authors: | Chen, J.L, Fountain, M.A, Disney, M.D. | | Deposit date: | 2019-12-16 | | Release date: | 2020-05-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design, Optimization, and Study of Small Molecules That Target Tau Pre-mRNA and Affect Splicing.
J.Am.Chem.Soc., 142, 2020
|
|
6VA2
 
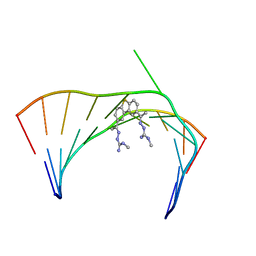 | | Solution Structure of the Tau pre-mRNA Exon 10 Splicing Regulatory Element Bound to MH5 | | Descriptor: | (2E,2'E)-2,2'-{dibenzo[b,d]thiene-2,8-diyldi[(1E)eth-1-yl-1-ylidene]}bis(N-methylhydrazine-1-carboximidamide), RNA (5'-R(*CP*AP*CP*AP*CP*GP*UP*CP*GP*G)-3'), RNA (5'-R(*CP*CP*GP*GP*CP*AP*GP*UP*GP*UP*G)-3') | | Authors: | Chen, J.L, Fountain, M.A, Disney, M.D. | | Deposit date: | 2019-12-16 | | Release date: | 2020-05-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Design, Optimization, and Study of Small Molecules That Target Tau Pre-mRNA and Affect Splicing.
J.Am.Chem.Soc., 142, 2020
|
|
6ICD
 
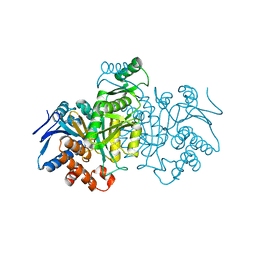 | | REGULATION OF AN ENZYME BY PHOSPHORYLATION AT THE ACTIVE SITE | | Descriptor: | ISOCITRATE DEHYDROGENASE | | Authors: | Hurley, J.H, Dean, A.M, Sohl, J.L, Koshlandjunior, D.E, Stroud, R.M. | | Deposit date: | 1990-05-30 | | Release date: | 1991-10-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Regulation of an enzyme by phosphorylation at the active site.
Science, 249, 1990
|
|
6VA1
 
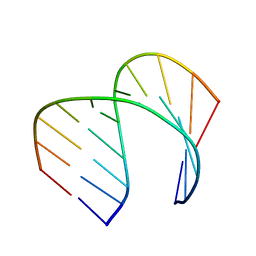 | | Solution Structure of the Tau pre-mRNA Exon 10 Splicing Regulatory Element | | Descriptor: | RNA (5'-R(*CP*AP*CP*AP*CP*GP*UP*CP*GP*G)-3'), RNA (5'-R(*CP*CP*GP*GP*CP*AP*GP*UP*GP*UP*G)-3') | | Authors: | Chen, J.L, Fountain, M.A, Disney, M.D. | | Deposit date: | 2019-12-16 | | Release date: | 2020-05-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Design, Optimization, and Study of Small Molecules That Target Tau Pre-mRNA and Affect Splicing.
J.Am.Chem.Soc., 142, 2020
|
|
5C4Z
 
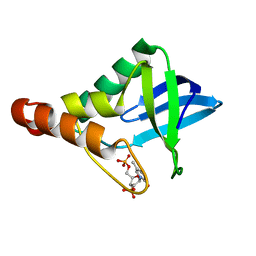 | |
