6YAJ
 
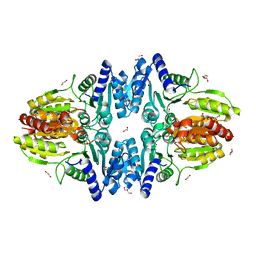 | | Split gene transketolase, inactive beta4 tetramer | | Descriptor: | 1,2-ETHANEDIOL, C-terminal chain of split transketolase from Carboxydothermus hydrogenoformans, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Isupov, M.N, Littlechild, J.A, James, P. | | Deposit date: | 2020-03-12 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A 'Split-Gene' Transketolase From the Hyper-Thermophilic Bacterium Carboxydothermus hydrogenoformans : Structure and Biochemical Characterization.
Front Microbiol, 11, 2020
|
|
5MQZ
 
 | | Archaeal branched-chain amino acid aminotransferase from Archaeoglobus fulgidus; holoform | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | James, P, Isupov, M.N, Sayer, C, Littlechild, J.A, Sutter, J.M, Schmidt, M, Schoenheit, P. | | Deposit date: | 2016-12-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Thermostable Branched-Chain Amino Acid Transaminases From the Archaea Geoglobus acetivorans and Archaeoglobus fulgidus : Biochemical and Structural Characterization.
Front Bioeng Biotechnol, 7, 2019
|
|
8P2M
 
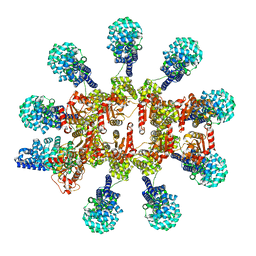 | | C. elegans TIR-1 protein. | | Descriptor: | NAD(+) hydrolase tir-1 | | Authors: | Isupov, M.N, Opatowsky, Y. | | Deposit date: | 2023-05-16 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structure-function analysis of ceTIR-1/hSARM1 explains the lack of Wallerian axonal degeneration in C. elegans.
Cell Rep, 42, 2023
|
|
8P2L
 
 | |
1QMV
 
 | | thioredoxin peroxidase B from red blood cells | | Descriptor: | PEROXIREDOXIN-2 | | Authors: | Isupov, M.N, Littlechild, J.A, Lebedev, A.A, Errington, N, Vagin, A.A, Schroder, E. | | Deposit date: | 1999-10-07 | | Release date: | 2000-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Decameric 2-Cys Peroxiredoxin from Human Erythrocytes at 1.7 A Resolution.
Structure, 8, 2000
|
|
4BQ0
 
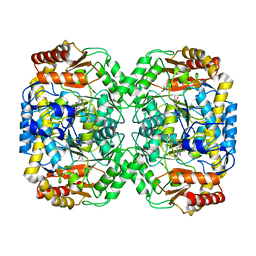 | | Pseudomonas aeruginosa beta-alanine:pyruvate aminotransferase holoenzyme without divalent cations on dimer-dimer interface | | Descriptor: | BETA-ALANINE--PYRUVATE TRANSAMINASE, CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Isupov, M.N, Lebedev, A.A, Westlake, A, Sayer, C, Littlechild, J.A. | | Deposit date: | 2013-05-29 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Space-Group and Origin Ambiguity in Macromolecular Structures with Pseudo-Symmetry and its Treatment with the Program Zanuda.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6T92
 
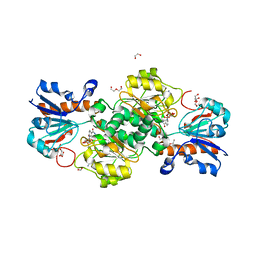 | | NAD+-dependent fungal formate dehydrogenase from Chaetomium thermophilum: A complex of N120C mutant protein with the reduced form of the cofactor NADH and the substrate formate at a secondary site. | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Isupov, M.N, Yelmazer, B, De Rose, S.A, Littlechild, J.A. | | Deposit date: | 2019-10-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural insights into the NAD + -dependent formate dehydrogenase mechanism revealed from the NADH complex and the formate NAD + ternary complex of the Chaetomium thermophilum enzyme.
J.Struct.Biol., 212, 2020
|
|
6T8Z
 
 | | NAD+-dependent fungal formate dehydrogenase from Chaetomium thermophilum: A ternary complex with the oxidised form of the cofactor NAD+ and the substrate formate both at a primary and secondary sites. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Isupov, M.N, Yelmazer, B, De Rose, S.A, Littlechild, J.A. | | Deposit date: | 2019-10-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structural insights into the NAD + -dependent formate dehydrogenase mechanism revealed from the NADH complex and the formate NAD + ternary complex of the Chaetomium thermophilum enzyme.
J.Struct.Biol., 212, 2020
|
|
6T94
 
 | | NAD+-dependent fungal formate dehydrogenase from Chaetomium thermophilum: A complex of N120C mutant protein with the reduced form of the cofactor NADH. | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Isupov, M.N, Yelmazer, B, De Rose, S.A, Littlechild, J.A. | | Deposit date: | 2019-10-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural insights into the NAD + -dependent formate dehydrogenase mechanism revealed from the NADH complex and the formate NAD + ternary complex of the Chaetomium thermophilum enzyme.
J.Struct.Biol., 212, 2020
|
|
6T8Y
 
 | | NAD+-dependent fungal formate dehydrogenase from Chaetomium thermophilum: A complex with the reduced form of the cofactor NADH and the substrate formate at a secondary site. | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Isupov, M.N, Yelmazer, B, De Rose, S.A, Littlechild, J.A. | | Deposit date: | 2019-10-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structural insights into the NAD + -dependent formate dehydrogenase mechanism revealed from the NADH complex and the formate NAD + ternary complex of the Chaetomium thermophilum enzyme.
J.Struct.Biol., 212, 2020
|
|
7PNB
 
 | | Sulfolobus acidocaldarius 0406 filament. | | Descriptor: | 6-deoxy-6-sulfo-beta-D-glucopyranose-(1-3)-[alpha-D-mannopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sulfolobus acidocaldarius 0406 filament., beta-D-glucopyranose-(1-4)-6-deoxy-6-sulfo-beta-D-glucopyranose-(1-3)-[alpha-D-mannopyranose-(1-4)][alpha-D-mannopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Isupov, M.N, Gaines, M, Daum, B. | | Deposit date: | 2021-09-06 | | Release date: | 2022-09-14 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Electron cryo-microscopy reveals the structure of the archaeal thread filament.
Nat Commun, 13, 2022
|
|
7C4D
 
 | | Marine microorganism esterase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Putative esterase, ... | | Authors: | Zhu, C.H, Wu, Y.K, Isupov, M.N. | | Deposit date: | 2020-05-16 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Insights into a Novel Esterase from the East Pacific Rise and Its Improved Thermostability by a Semirational Design.
J.Agric.Food Chem., 69, 2021
|
|
4UHC
 
 | | Structural studies of a thermophilic esterase from Thermogutta terrifontis (Native) | | Descriptor: | CHLORIDE ION, ESTERASE | | Authors: | Sayer, C, Isupov, M.N, Bonch-Osmolovskaya, E, Littlechild, J.A. | | Deposit date: | 2015-03-24 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Structural Studies of a Thermophilic Esterase from a New Planctomycetes Species, Thermogutta Terrifontis.
FEBS J., 282, 2015
|
|
4UHE
 
 | | Structural studies of a thermophilic esterase from Thermogutta terrifontis (malate bound) | | Descriptor: | CHLORIDE ION, D-MALATE, ESTERASE, ... | | Authors: | Sayer, C, Isupov, M.N, Bonch-Osmolovskaya, E, Littlechild, J.A. | | Deposit date: | 2015-03-24 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structural Studies of a Thermophilic Esterase from a New Planctomycetes Species, Thermogutta Terrifontis.
FEBS J., 282, 2015
|
|
6KXT
 
 | | BON1-C2B | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Protein BONZAI 1, SULFATE ION | | Authors: | Wang, Q.C, Jiang, M.Q, Isupov, M.N, Sun, L.F, Wu, Y.K. | | Deposit date: | 2019-09-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of an Arabidopsis Copine providing insights into this protein family
To be published
|
|
6KXU
 
 | | BON1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wang, Q.C, Jiang, M.Q, Isupov, M.N, Sun, L.F, Wu, Y.K. | | Deposit date: | 2019-09-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Crystal Structure of an Arabidopsis Copine providing insights into this protein family
To be published
|
|
6KXK
 
 | | BON1 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wang, Q.C, Jiang, M.Q, Isupov, M.N, Sun, L.F, Wu, Y.K. | | Deposit date: | 2019-09-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of an Arabidopsis Copine providing insights into this protein family
To be published
|
|
1H2B
 
 | | Crystal Structure of the Alcohol Dehydrogenase from the Hyperthermophilic Archaeon Aeropyrum pernix at 1.65A Resolution | | Descriptor: | ALCOHOL DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE (ACIDIC FORM), OCTANOIC ACID (CAPRYLIC ACID), ... | | Authors: | Guy, J.E, Isupov, M.N, Littlechild, J.A. | | Deposit date: | 2002-08-02 | | Release date: | 2003-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The structure of an alcohol dehydrogenase from the hyperthermophilic archaeon Aeropyrum pernix.
J.Mol.Biol., 331, 2003
|
|
5FRD
 
 | | Structure of a thermophilic esterase | | Descriptor: | CARBOXYLESTERASE (EST-2), CHLORIDE ION, CITRATE ANION, ... | | Authors: | Sayer, C, Finnigan, W, Isupov, M.N, Levisson, M, Kengen, S.W.M, van der Oost, J, Harmer, N, Littlechild, J.A. | | Deposit date: | 2015-12-17 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Biochemical Characterisation of Archaeoglobus Fulgidus Esterase Reveals a Bound Coa Molecule in the Vicinity of the Active Site.
Sci.Rep., 6, 2016
|
|
5H3V
 
 | | Crystal structure of a Type IV Secretion System Component CagX in Helicobacter pylori | | Descriptor: | Cag8, DI(HYDROXYETHYL)ETHER, ISOPROPYL ALCOHOL | | Authors: | Zhang, J, Wu, Y, Zhao, Y, Sun, L, Keegan, R.M, Liu, Y, Isupov, M.N. | | Deposit date: | 2016-10-27 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the type IV secretion system component CagX from Helicobacter pylori
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5I6J
 
 | |
8EDD
 
 | | Staphylococcus aureus endonuclease IV Y33F mutant | | Descriptor: | CHLORIDE ION, FE (III) ION, PHOSPHATE ION, ... | | Authors: | Saper, M.A, Kirillov, S, Isupov, M.N, Wiener, R, Rouvinski, A. | | Deposit date: | 2022-09-04 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Octahedrally coordinated iron in the catalytic site of endonuclease IV from Staphylococcus aureus
To Be Published
|
|
5I7D
 
 | |
5I6R
 
 | | Crystal Structure of srGAP2 F-BARx WT Form-1 | | Descriptor: | ACETATE ION, D-MALATE, SLIT-ROBO Rho GTPase-activating protein 2, ... | | Authors: | Sporny, M, Guez-Haddad, J, Isupov, M.N, Opatowsky, Y. | | Deposit date: | 2016-02-16 | | Release date: | 2017-08-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for srGAP2 Membrane Interactions, and Antagonism by the Human Specific Paralog srGAP2C
To Be Published
|
|
3SS9
 
 | | Crystal structure of holo D-serine dehydratase from Escherichia coli at 1.97 A resolution | | Descriptor: | D-serine dehydratase, POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Urusova, D.V, Isupov, M.N, Antonyuk, S.V, Kachalova, G.S, Vagin, A.A, Lebedev, A.A, Bourenkov, G.P, Dauter, Z, Bartunik, H.D, Melik-Adamyan, W.R, Mueller, T.D, Schnackerz, K.D. | | Deposit date: | 2011-07-08 | | Release date: | 2012-01-18 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of D-serine dehydratase from Escherichia coli.
Biochim.Biophys.Acta, 1824, 2011
|
|
