7ZYK
 
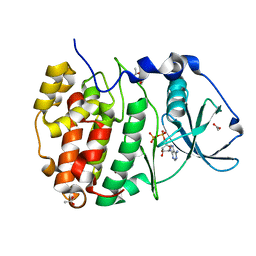 | | Compound 9 Bound to CK2alpha | | Descriptor: | 2-(5-bromanyl-6-chloranyl-1~{H}-indol-3-yl)ethanenitrile, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Brear, P, Hyvonen, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | A fragment-based approach leading to the discovery of inhibitors of CK2 alpha with a novel mechanism of action.
Rsc Med Chem, 13, 2022
|
|
8AEM
 
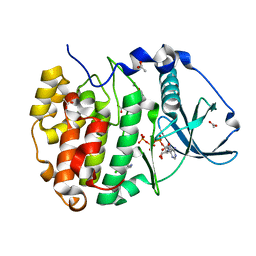 | | Structure of Compound 13 bound to CK2alpha | | Descriptor: | 2-(5-chloranyl-1H-indol-3-yl)ethanenitrile, ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Brear, P, Fusco, C, Atkinson, E, Iegre, J, Francis-Newton, N, Venkotaraman, A, Spring, D, Hyvonen, M. | | Deposit date: | 2022-07-13 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A fragment-based approach leading to the discovery of inhibitors of CK2 alpha with a novel mechanism of action.
Rsc Med Chem, 13, 2022
|
|
8AE7
 
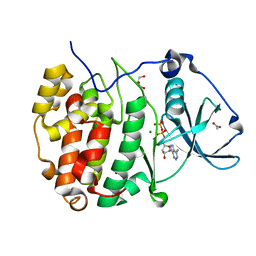 | | The strucuture of Compound 15 bound to CK2alpha | | Descriptor: | 2-[5,6-bis(bromanyl)-1H-indazol-3-yl]ethanenitrile, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Brear, P, De Fusco, C, Atkinson, E, Frances, N, Iegre, J, Venkitaraman, A, Hyvonen, M, Spring, D. | | Deposit date: | 2022-07-12 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | A fragment-based approach leading to the discovery of inhibitors of CK2 alpha with a novel mechanism of action.
Rsc Med Chem, 13, 2022
|
|
8BE1
 
 | | SARS-CoV-2 RBD in complex with a Fab fragment of a neutralising antibody mRBD2 | | Descriptor: | Antibody heavy chain, Antibody light chain, SULFATE ION, ... | | Authors: | Lulla, A, Brear, P, Fischer, K, Hollfelder, F, Hyvonen, M. | | Deposit date: | 2022-10-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Microfluidics-enabled fluorescence-activated cell sorting of single pathogen-specific antibody secreting cells for the rapid discovery of monoclonal antibodies
Biorxiv, 2023
|
|
8C3J
 
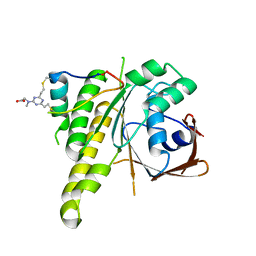 | | Stapled peptide SP2 in complex with humanised RadA mutant HumRadA22 | | Descriptor: | 2-[(4,6-diethyl-1,3,5-triazin-2-yl)-methyl-amino]ethanoic acid, Breast cancer type 2 susceptibility protein, DNA repair and recombination protein RadA | | Authors: | Pantelejevs, T, Hyvonen, M. | | Deposit date: | 2022-12-26 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | A recombinant approach for stapled peptide discovery yields inhibitors of the RAD51 recombinase.
Chem Sci, 14, 2023
|
|
4A6X
 
 | | RadA C-terminal ATPase domain from Pyrococcus furiosus bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA REPAIR AND RECOMBINATION PROTEIN RADA, MAGNESIUM ION | | Authors: | Marsh, M.E, Ehebauer, M.T, Scott, D, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2011-11-10 | | Release date: | 2012-11-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.548 Å) | | Cite: | ATP Half-Sites in Rada and Rad51 Recombinases Bind Nucleotides
FEBS Open Bio, 6, 2016
|
|
4A6P
 
 | | RadA C-terminal ATPase domain from Pyrococcus furiosus | | Descriptor: | DNA REPAIR AND RECOMBINATION PROTEIN RADA, PHOSPHATE ION | | Authors: | Marsh, M.E, Ehebauer, M.T, Scott, D, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2011-11-08 | | Release date: | 2012-11-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | ATP Half-Sites in Rada and Rad51 Recombinases Bind Nucleotides
FEBS Open Bio, 6, 2016
|
|
4B2P
 
 | | RadA C-terminal ATPase domain from Pyrococcus furiosus bound to GTP | | Descriptor: | DNA REPAIR AND RECOMBINATION PROTEIN RADA, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Marsh, M.E, Ehebauer, M.T, Scott, D, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2012-07-17 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | ATP Half-Sites in Rada and Rad51 Recombinases Bind Nucleotides
FEBS Open Bio, 6, 2016
|
|
5AFG
 
 | | Structure of the Stapled Peptide Bound to Mdm2 | | Descriptor: | 1,8-DIETHYL-1,8-DIHYDRODIBENZO[3,4:7,8][1,2,3]TRIAZOLO[4',5':5,6]CYCLOOCTA[1,2-D][1,2,3]TRIAZOLE, E3 UBIQUITIN-PROTEIN LIGASE MDM2, STAPLED PEPTIDE | | Authors: | Lau, Y.H, Wu, Y, Rossmann, M, de Andrade, P, Tan, Y.S, McKenzie, G.J, Venkitaraman, A.R, Hyvonen, M, Spring, D.R. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Double Strain-Promoted Macrocyclization for the Rapid Selection of Cell-Active Stapled Peptides.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5AEJ
 
 | | Crystal structure of human Gremlin-1 | | Descriptor: | GREMLIN-1, SULFATE ION | | Authors: | Kisonaite, M, Hyvonen, M. | | Deposit date: | 2015-08-31 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Structure of Gremlin-1 and Analysis of its Interaction with Bmp-2.
Biochem.J., 473, 2016
|
|
5BXO
 
 | | Human Tankyrase-2 in Complex with Macrocyclised Extended Peptide cp4n2m3 | | Descriptor: | 4,4'-propane-1,3-diylbis(1-methyl-1H-1,2,3-triazole), DIMETHYL SULFOXIDE, SULFATE ION, ... | | Authors: | Xu, W, Fischer, G, Hyvonen, M, Itzhaki, L. | | Deposit date: | 2015-06-09 | | Release date: | 2016-06-29 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (1.334 Å) | | Cite: | Macrocyclized Extended Peptides: Inhibiting the Substrate-Recognition Domain of Tankyrase.
J.Am.Chem.Soc., 139, 2017
|
|
5OBR
 
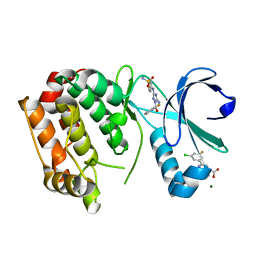 | | Aurora A kinase in complex with 2-(3-chloro-5-fluorophenyl)quinoline-4-carboxylic acid and JNJ-7706621 | | Descriptor: | 2-(3-chloranyl-5-fluoranyl-phenyl)quinoline-4-carboxylic acid, 4-({5-amino-1-[(2,6-difluorophenyl)carbonyl]-1H-1,2,4-triazol-3-yl}amino)benzenesulfonamide, Aurora kinase A, ... | | Authors: | Rossmann, M, Janecek, M, Hyvonen, M. | | Deposit date: | 2017-06-29 | | Release date: | 2017-08-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Computationally-guided optimization of small-molecule inhibitors of the Aurora A kinase-TPX2 protein-protein interaction.
Chem. Commun. (Camb.), 53, 2017
|
|
5OBJ
 
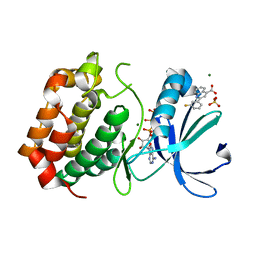 | | Aurora A kinase in complex with 2-(3-fluorophenyl)quinoline-4-carboxylic acid and ATP | | Descriptor: | 2-(3-fluorophenyl)quinoline-4-carboxylic acid, ADENOSINE-5'-TRIPHOSPHATE, Aurora kinase A, ... | | Authors: | Rossmann, M, Janecek, M, Hyvonen, M. | | Deposit date: | 2017-06-28 | | Release date: | 2017-08-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Computationally-guided optimization of small-molecule inhibitors of the Aurora A kinase-TPX2 protein-protein interaction.
Chem. Commun. (Camb.), 53, 2017
|
|
4ZI5
 
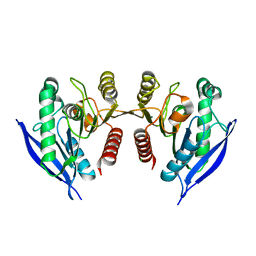 | | Crystal Structure of Dienelactone Hydrolase-like Promiscuous Phospotriesterase P91 from Metagenomic Libraries | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, P91 | | Authors: | Colin, P.-Y, Fischer, G, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2015-04-27 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Ultrahigh-throughput discovery of promiscuous enzymes by picodroplet functional metagenomics.
Nat Commun, 6, 2015
|
|
5AJ9
 
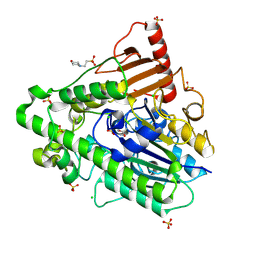 | | G7 mutant of PAS, arylsulfatase from Pseudomonas Aeruginosa | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ARYLSULFATASE, CALCIUM ION, ... | | Authors: | Miton, C.M, Fischer, G, Jonas, S, Mohammed, M.F, Loo, B.v, Kintses, B, Hyvonen, M, Tokuriki, N, Hollfelder, F. | | Deposit date: | 2015-02-20 | | Release date: | 2016-03-16 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolutionary repurposing of a sulfatase: A new Michaelis complex leads to efficient transition state charge offset.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5BXU
 
 | | Human Tankyrase-2 in Complex with Macrocyclised Extended Peptide cp4n4m5 | | Descriptor: | 4,4'-pentane-1,5-diylbis(1-propyl-1H-1,2,3-triazole), CHLORIDE ION, Tankyrase-2, ... | | Authors: | Xu, W, Fischer, G, Hyvonen, M, Itzhaki, L. | | Deposit date: | 2015-06-09 | | Release date: | 2016-06-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Human Tankyrase-2 in Complex with Extended Stapled Peptide sp4n4m5
to be published
|
|
5CLP
 
 | | Crystal Structure of CK2alpha with 3,4-dichlorophenethylamine bound | | Descriptor: | 2-(3,4-dichlorophenyl)ethanamine, ACETATE ION, Casein kinase II subunit alpha | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-16 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.684 Å) | | Cite: | Specific inhibition of CK2 alpha from an anchor outside the active site.
Chem Sci, 7, 2016
|
|
5CSV
 
 | | Crystal Structure of CK2alpha with Compound 6 bound | | Descriptor: | 3-AMINOBENZOIC ACID, ACETATE ION, Casein kinase II subunit alpha | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-23 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.375 Å) | | Cite: | Specific inhibition of CK2 alpha from an anchor outside the active site.
Chem Sci, 7, 2016
|
|
5CU4
 
 | | Crystal structure of CK2alpha bound to CAM4066 | | Descriptor: | ACETATE ION, Casein kinase II subunit alpha, N-[(2-chlorobiphenyl-4-yl)methyl]-beta-alanyl-N-(3-carboxyphenyl)-beta-alaninamide | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-24 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Specific inhibition of CK2 alpha from an anchor outside the active site.
Chem Sci, 7, 2016
|
|
5CVG
 
 | | Crystal Structure of CK2alpha with a novel closed conformation of the aD loop | | Descriptor: | ACETATE ION, Casein kinase II subunit alpha | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-26 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Specific inhibition of CK2 alpha from an anchor outside the active site.
Chem Sci, 7, 2016
|
|
5CU0
 
 | | Crystal structure of CK2alpha with 2-hydroxy-5-methylbenzoic acid and N-(3-(3-chloro-4-(phenyl)benzylamino)propyl)acetamide bound | | Descriptor: | 2-hydroxy-5-methylbenzoic acid, ACETATE ION, Casein kinase II subunit alpha, ... | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-24 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | A fragment-based approach leading to the discovery of a novel binding site and the selective CK2 inhibitor CAM4066.
Bioorg. Med. Chem., 25, 2017
|
|
5CT0
 
 | | Crystal structure of CK2alpha with 3-(3-chloro-4-(phenyl)benzylamino)propan-1-ol bound | | Descriptor: | 3-{[(2-chlorobiphenyl-4-yl)methyl]amino}propan-1-ol, ACETATE ION, Casein kinase II subunit alpha | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-23 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.008 Å) | | Cite: | A fragment-based approach leading to the discovery of a novel binding site and the selective CK2 inhibitor CAM4066.
Bioorg. Med. Chem., 25, 2017
|
|
5CSP
 
 | | Crystal Structure of CK2alpha with Compound 5 bound | | Descriptor: | 2-hydroxy-5-methylbenzoic acid, ACETATE ION, Casein kinase II subunit alpha | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-23 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Specific inhibition of CK2 alpha from an anchor outside the active site.
Chem Sci, 7, 2016
|
|
5CS6
 
 | | Crystal Structure of CK2alpha with Compound 3 bound | | Descriptor: | 1-(3-chloro-4-propoxyphenyl)methanamine, ACETATE ION, Casein kinase II subunit alpha, ... | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-23 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Specific inhibition of CK2 alpha from an anchor outside the active site.
Chem Sci, 7, 2016
|
|
5CU6
 
 | | Crystal Structure of CK2alpha | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, Casein kinase II subunit alpha | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-24 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Specific inhibition of CK2 alpha from an anchor outside the active site.
Chem Sci, 7, 2016
|
|
