7FS8
 
 | | Structure of liver pyruvate kinase in complex with allosteric modulator 21 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 4-[4-(3-aminobenzene-1-sulfonyl)piperazine-1-sulfonyl]benzene-1,2-diol, MAGNESIUM ION, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
7FS9
 
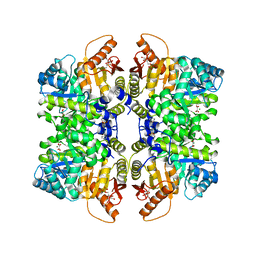 | | Structure of liver pyruvate kinase in complex with allosteric modulator 22 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 4-[4-(4-aminobenzene-1-sulfonyl)piperazine-1-sulfonyl]benzene-1,2-diol, MAGNESIUM ION, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Method: | X-RAY DIFFRACTION (1.721 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
5HLZ
 
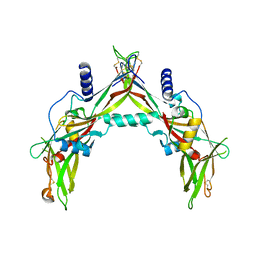 | |
5HLY
 
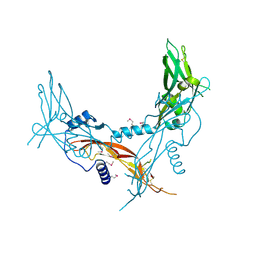 | |
6Q4Q
 
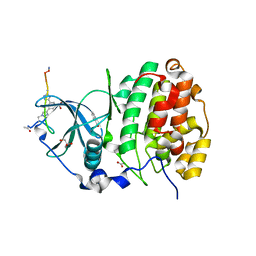 | | The Crystal structure of CK2a bound to P2-C4 | | Descriptor: | 3,5-bis(1-methyl-1,2,3-triazol-4-yl)benzoic acid, ACETATE ION, BENZOIC ACID, ... | | Authors: | Brear, P, Iegre, J, Baker, D, Tan, Y, Sore, H, Donovan, D, Spring, D, Chandra, V, Hyvonen, M. | | Deposit date: | 2018-12-06 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Efficient development of stable and highly functionalised peptides targeting the CK2 alpha /CK2 beta protein-protein interaction.
Chem Sci, 10, 2019
|
|
6Q38
 
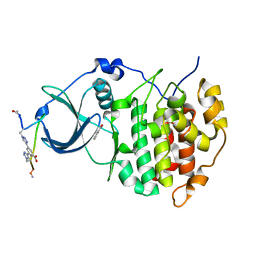 | | The Crystal structure of CK2a bound to P1-C4 | | Descriptor: | 3,5-bis(1-methyl-1,2,3-triazol-4-yl)benzoic acid, BENZOIC ACID, Casein kinase II subunit alpha, ... | | Authors: | Brear, P, Iegre, J, Baker, D, Tan, Y, Sore, H, Donovan, D, Spring, D, Chandra, V, Hyvonen, M. | | Deposit date: | 2018-12-03 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Efficient development of stable and highly functionalised peptides targeting the CK2 alpha /CK2 beta protein-protein interaction.
Chem Sci, 10, 2019
|
|
6RK1
 
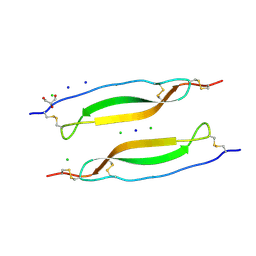 | | Crystal structure of TSP1 domain from CCN3 | | Descriptor: | CCN family member 3, CHLORIDE ION, GLYCEROL, ... | | Authors: | Xu, E.-R, Hyvonen, M. | | Deposit date: | 2019-04-29 | | Release date: | 2019-10-02 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | The thrombospondin module 1 domain of the matricellular protein CCN3 shows an atypical disulfide pattern and incomplete CWR layers.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
3P2Z
 
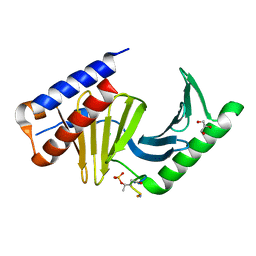 | | Polo-like kinase I Polo-box domain in complex with PLHSpTA phosphopeptide from PBIP1 | | Descriptor: | GLYCEROL, Serine/threonine-protein kinase PLK1, phosphopeptide | | Authors: | Sledz, P, Stubbs, C.J, Hyvonen, M, Abell, C. | | Deposit date: | 2010-10-04 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | From crystal packing to molecular recognition: prediction and discovery of a binding site on the surface of polo-like kinase 1
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
3P36
 
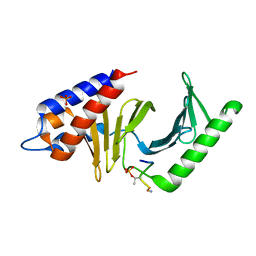 | | Polo-like kinase I Polo-box domain in complex with DPPLHSpTA phosphopeptide from PBIP1 | | Descriptor: | GLYCEROL, PHOSPHATE ION, Serine/threonine-protein kinase PLK1, ... | | Authors: | Sledz, P, Stubbs, C.J, Hyvonen, M, Abell, C. | | Deposit date: | 2010-10-04 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | From crystal packing to molecular recognition: prediction and discovery of a binding site on the surface of polo-like kinase 1
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
3P2W
 
 | | Unliganded form of Polo-like kinase I Polo-box domain | | Descriptor: | GLYCEROL, SULFATE ION, Serine/threonine-protein kinase PLK1 | | Authors: | Sledz, P, Hyvonen, M, Abell, C. | | Deposit date: | 2010-10-04 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | From crystal packing to molecular recognition: prediction and discovery of a binding site on the surface of polo-like kinase 1
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
3P34
 
 | | Polo-like kinase I Polo-box domain in complex with MQSpTPL phosphopeptide | | Descriptor: | GLYCEROL, SULFATE ION, Serine/threonine-protein kinase PLK1, ... | | Authors: | Sledz, P, Hyvonen, M, Abell, C. | | Deposit date: | 2010-10-04 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | From crystal packing to molecular recognition: prediction and discovery of a binding site on the surface of polo-like kinase 1
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
3P35
 
 | |
3P37
 
 | |
3Q1I
 
 | | Polo-like kinase I Polo-box domain in complex with FMPPPMSpSM phosphopeptide from TCERG1 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | Authors: | Sledz, P, Hyvonen, M, Abell, C. | | Deposit date: | 2010-12-17 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | From crystal packing to molecular recognition: prediction and discovery of a binding site on the surface of polo-like kinase 1
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
6XTW
 
 | | HumRadA33F in complex with peptidic inhibitor 6 | | Descriptor: | DNA repair and recombination protein RadA, SULFATE ION, ~{N}-[2-[(2~{S})-2-[[(1~{S})-1-(4-methoxyphenyl)ethyl]carbamoyl]pyrrolidin-1-yl]-2-oxidanylidene-ethyl]quinoline-2-carboxamide | | Authors: | Fischer, G, Marsh, M.E, Scott, D.E, Coyne, A.G, Skidmore, J, Abell, C, Hyvonen, M. | | Deposit date: | 2020-01-16 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death.
Cell Chem Biol, 28, 2021
|
|
7QEA
 
 | | Crystal structure of fluorescein-di-Beta-D-glucuronide bound to a mutant of SN243 (D415A) | | Descriptor: | (2~{S},3~{S},4~{S},5~{R},6~{S})-3,4,5-tris(oxidanyl)-6-[(1~{R})-6'-oxidanyl-3-oxidanylidene-spiro[2-benzofuran-1,9'-xanthene]-3'-yl]oxy-oxane-2-carboxylic acid, ACETATE ION, SN243, ... | | Authors: | Neun, S, Brear, P, Campbell, E, Omari, K, Wagner, O, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2021-12-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Functional metagenomic screening identifies an unexpected beta-glucuronidase.
Nat.Chem.Biol., 18, 2022
|
|
7QEF
 
 | | Crystal structure of para-nitrophenyl-Beta-D-glucuronide bound to a mutant of SN243 (D415A) | | Descriptor: | 4-nitrophenyl beta-D-glucopyranosiduronic acid, ACETATE ION, SN243, ... | | Authors: | Neun, S, Brear, P, Campbell, E, Omari, K, Wagner, O, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2021-12-02 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Functional metagenomic screening identifies an unexpected beta-glucuronidase.
Nat.Chem.Biol., 18, 2022
|
|
7QE1
 
 | | Crystal structure of apo SN243 | | Descriptor: | SN243, ZINC ION | | Authors: | Neun, S, Brear, P, Campbell, E, Omari, K, Wagner, O, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2021-12-01 | | Release date: | 2022-10-12 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Functional metagenomic screening identifies an unexpected beta-glucuronidase.
Nat.Chem.Biol., 18, 2022
|
|
7QE2
 
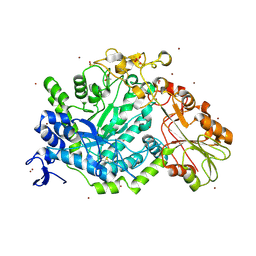 | | Crystal structure of D-glucuronic acid bound to SN243 | | Descriptor: | ACETATE ION, SN243, SULFATE ION, ... | | Authors: | Neun, S, Brear, P, Campbell, E, Omari, K, Wagner, O, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2021-12-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Functional metagenomic screening identifies an unexpected beta-glucuronidase.
Nat.Chem.Biol., 18, 2022
|
|
7QG4
 
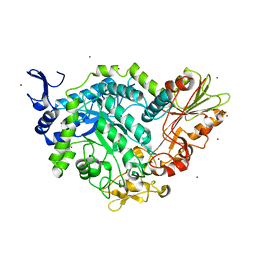 | | Apo crystal structure of a mutant of SN243 (D415N) | | Descriptor: | SN243, SULFATE ION, ZINC ION | | Authors: | Neun, S, Brear, P, Campbell, E, Omari, K, Wagner, O, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2021-12-07 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Functional metagenomic screening identifies an unexpected beta-glucuronidase.
Nat.Chem.Biol., 18, 2022
|
|
7QEE
 
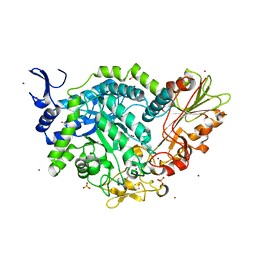 | | SN243 mutant D415N bound to para-nitrophenyl-Beta-D-glucuronide | | Descriptor: | 4-nitrophenyl beta-D-glucopyranosiduronic acid, SN243, SULFATE ION, ... | | Authors: | Neun, S, Brear, P, Campbell, E, Omari, K, Wagner, O, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2021-12-02 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.374 Å) | | Cite: | Functional metagenomic screening identifies an unexpected beta-glucuronidase.
Nat.Chem.Biol., 18, 2022
|
|
6YYK
 
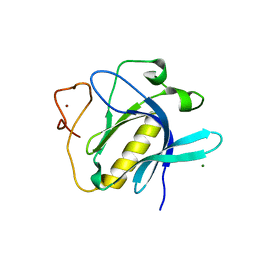 | | Crystal Structure of 1,5-dimethylindoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase mutant R28C | | Descriptor: | 1,5-dimethyl-3~{H}-indol-2-one, MAGNESIUM ION, Tyrosine-protein kinase BTK, ... | | Authors: | Brear, P, Wagstaff, J, Hyvonen, M. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Optimising crystallographic systems for structure-guided drug discovery
To Be Published
|
|
6YYF
 
 | | Crystal Structure of 5-chloroindoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase mutant R28C | | Descriptor: | 5-chloranyl-1,3-dihydroindol-2-one, MAGNESIUM ION, Tyrosine-protein kinase BTK, ... | | Authors: | Brear, P, Wagstaff, J, Hyvonen, M. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Optimising crystallographic systems for structure-guided drug discovery
To Be Published
|
|
6YYG
 
 | | Crystal Structure of 5-(trifluoromethoxy)indoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase mutant R28C | | Descriptor: | 5-(trifluoromethyloxy)-1,3-dihydroindol-2-one, MAGNESIUM ION, Tyrosine-protein kinase BTK, ... | | Authors: | Brear, P, Wagstaff, J, Hyvonen, M. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Optimising crystallographic systems for structure-guided drug discovery
To Be Published
|
|
7ZY8
 
 | | Crystal structure of compound 2 bound to CK2alpha | | Descriptor: | 3-[3,5-bis(chloranyl)phenyl]propan-1-amine, ACETATE ION, Casein kinase II subunit alpha, ... | | Authors: | Brear, P, Fusco, C, Atkinson, E, Rossmann, M, Francis, N, Iegre, J, Hyvonen, M, Spring, D. | | Deposit date: | 2022-05-24 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A fragment-based approach leading to the discovery of inhibitors of CK2 alpha with a novel mechanism of action.
Rsc Med Chem, 13, 2022
|
|
