1B55
 
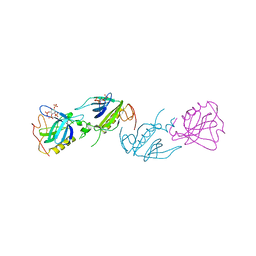 | | PH DOMAIN FROM BRUTON'S TYROSINE KINASE IN COMPLEX WITH INOSITOL 1,3,4,5-TETRAKISPHOSPHATE | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, TYROSINE-PROTEIN KINASE BTK, ZINC ION | | Authors: | Djinovic Carugo, K, Baraldi, E, Hyvoenen, M, Lo Surdo, P, Riley, A.M, Potter, B.V.L, O'Brien, R, Ladbury, J.E, Saraste, M. | | Deposit date: | 1999-01-12 | | Release date: | 1999-06-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the PH domain from Bruton's tyrosine kinase in complex with inositol 1,3,4,5-tetrakisphosphate.
Structure Fold.Des., 7, 1999
|
|
1BWN
 
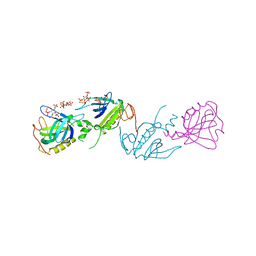 | | PH DOMAIN AND BTK MOTIF FROM BRUTON'S TYROSINE KINASE MUTANT E41K IN COMPLEX WITH INS(1,3,4,5)P4 | | Descriptor: | BRUTON'S TYROSINE KINASE, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, ZINC ION | | Authors: | Djinovic Carugo, K, Baraldi, E, Hyvoenen, M, Lo Surdo, P, Riley, A, Potter, B, Saraste, M. | | Deposit date: | 1998-09-25 | | Release date: | 1999-06-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the PH domain from Bruton's tyrosine kinase in complex with inositol 1,3,4,5-tetrakisphosphate.
Structure Fold.Des., 7, 1999
|
|
6XUF
 
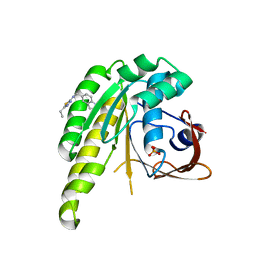 | | HumRadA1 in complex with 5-Ethyl-N-(1H-indol-5-ylmethyl)-1,3,4-thiadiazol-2-amine in P21 | | Descriptor: | 5-Ethyl-N-(1H-indol-5-ylmethyl)-1,3,4-thiadiazol-2-amine, DNA repair and recombination protein RadA, PHOSPHATE ION | | Authors: | Marsh, M.E, Scott, D.E, Hyvonen, M.E. | | Deposit date: | 2020-01-19 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.241 Å) | | Cite: | Optimising crystallographic systems for structure-guided drug discovery
To be published
|
|
1MPH
 
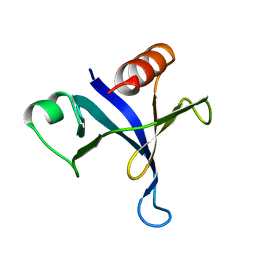 | | PLECKSTRIN HOMOLOGY DOMAIN FROM MOUSE BETA-SPECTRIN, NMR, 50 STRUCTURES | | Descriptor: | BETA SPECTRIN | | Authors: | Nilges, M, Macias, M.J, O'Donoghue, S.I, Oschkinat, H. | | Deposit date: | 1997-04-23 | | Release date: | 1997-06-16 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Automated NOESY interpretation with ambiguous distance restraints: the refined NMR solution structure of the pleckstrin homology domain from beta-spectrin.
J.Mol.Biol., 269, 1997
|
|
6Y6Z
 
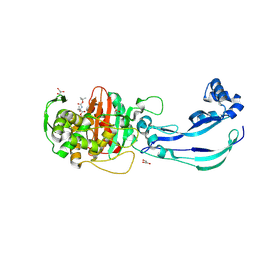 | | Structure of Pseudomonas aeruginosa Penicillin-Binding Protein 3 (PBP3) in complex with Compound 1 | | Descriptor: | GLYCEROL, Peptidoglycan D,D-transpeptidase FtsI, ~{tert}-butyl ~{N}-[(2~{S})-2-methyl-4-oxidanyl-1-oxidanylidene-pent-4-en-2-yl]carbamate | | Authors: | Newman, H, Bellini, D, Dowson, C.G. | | Deposit date: | 2020-02-27 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Demonstration of the utility of DOS-derived fragment libraries for rapid hit derivatisation in a multidirectional fashion.
Chem Sci, 11, 2020
|
|
6Y6U
 
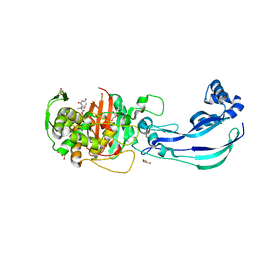 | | Structure of Pseudomonas aeruginosa Penicillin-Binding Protein 3 (PBP3) in complex with Compound 6 | | Descriptor: | 2-(4-hydroxyphenyl)-~{N}-[(2~{S})-2-methyl-4-oxidanyl-1-oxidanylidene-pent-4-en-2-yl]ethanamide, GLYCEROL, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Newman, H, Bellini, D, Dowson, C.G. | | Deposit date: | 2020-02-27 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Demonstration of the utility of DOS-derived fragment libraries for rapid hit derivatisation in a multidirectional fashion
Chem Sci, 11, 2020
|
|
6FVG
 
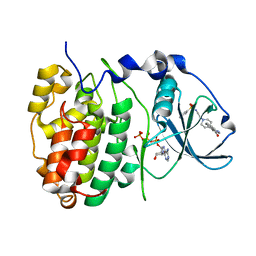 | | The Structure of CK2alpha with CCh507 bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Casein kinase II subunit alpha, [1-[2-(phenylsulfonylamino)ethyl]piperidin-4-yl]methyl 1~{H}-indole-3-carboxylate | | Authors: | Brear, P, Prudent, R, Laudet, B, Filhol, O, Cochet, C, Sautel, C, Moucadel, V, Bestgen, B, Engel, M, Ettaoussi, M, Lomberget, T, Le Borgne, M, Kufareva, I, Abagyan, R, Hyvonen, M. | | Deposit date: | 2018-03-02 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of holoenzyme-disrupting chemicals as substrate-selective CK2 inhibitors.
Sci Rep, 9, 2019
|
|
6FVF
 
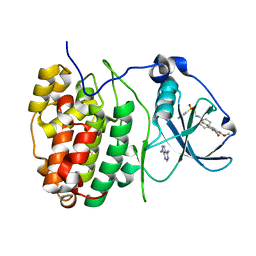 | | The Structure of CK2alpha with CCh503 bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Casein kinase II subunit alpha, [1-[2-(phenylsulfonylamino)ethyl]piperidin-4-yl]methyl 5-fluoranyl-2-methoxy-1~{H}-indole-3-carboxylate | | Authors: | Brear, P, Prudent, R, Laudet, B, Filhol, O, Cochet, C, Sautel, C, Moucadel, V, Bestgen, B, Engel, M, Ettaoussi, M, Lomberget, T, Le Borgne, M, Kufareva, I, Abagyan, R, Hyvonen, M. | | Deposit date: | 2018-03-02 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery of holoenzyme-disrupting chemicals as substrate-selective CK2 inhibitors.
Sci Rep, 9, 2019
|
|
6Y4Q
 
 | | Structure of a stapled peptide bound to MDM2 | | Descriptor: | ACE-LEU-THR-PHE-GLY-GLU-TYR-TRP-ALA-GLN-LEU-ALA-SER, E3 ubiquitin-protein ligase Mdm2, ~{N}-[(1-ethyl-1,2,3-triazol-4-yl)methyl]-~{N},5-dimethyl-4-[2-[2-methyl-5-[methyl-[(1-propyl-1,2,3-triazol-4-yl)methyl]carbamoyl]thiophen-3-yl]cyclopenten-1-yl]thiophene-2-carboxamide | | Authors: | Pantelejevs, T, Bakanovych, I. | | Deposit date: | 2020-02-22 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Diarylethene moiety as an enthalpy-entropy switch: photoisomerizable stapled peptides for modulating p53/MDM2 interaction.
Org.Biomol.Chem., 18, 2020
|
|
5UM2
 
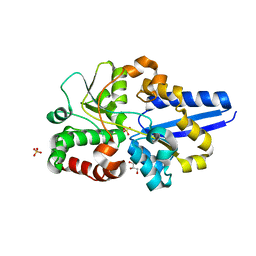 | |
5QUE
 
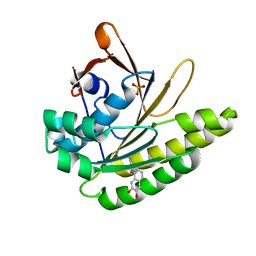 | |
5QUF
 
 | |
5QUB
 
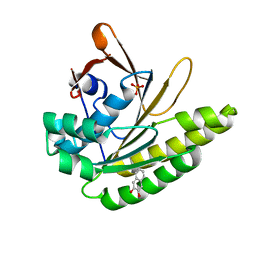 | |
5QUC
 
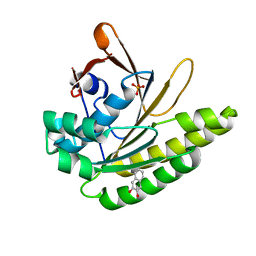 | |
5QUH
 
 | |
5QUD
 
 | |
5QUG
 
 | |
5QUO
 
 | |
5QUI
 
 | |
5QUQ
 
 | |
5QUP
 
 | |
5QUJ
 
 | |
5QUK
 
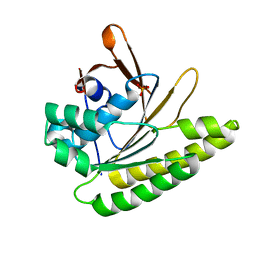 | | Structure of unliganded HumRadA1.2 | | Descriptor: | PHOSPHATE ION, RadA | | Authors: | Marsh, M, Hyvonen, M. | | Deposit date: | 2020-01-27 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Optimisation of crystal forms for structure-guided drug discovery
To be published
|
|
5QUN
 
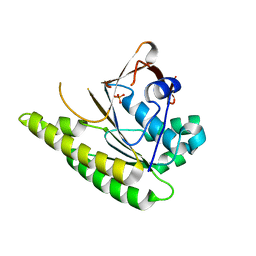 | | Structure of unliganded HumRadA1.6 | | Descriptor: | PHOSPHATE ION, RadA | | Authors: | Marsh, M, Hyvonen, M. | | Deposit date: | 2020-01-27 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Optimisation of crystal forms for structure-guided drug discovery
To be published
|
|
5QUM
 
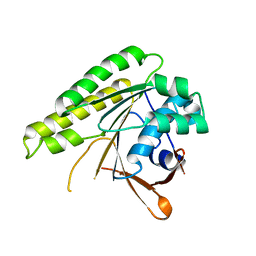 | | Structure of unliganded HumRadA1.4 | | Descriptor: | RadA | | Authors: | Marsh, M, Hyvonen, M. | | Deposit date: | 2020-01-27 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Optimisation of crystal forms for structure-guided drug discovery
To be published
|
|
