6IB7
 
 | | Structure of wild type SuhB | | Descriptor: | GLYCEROL, Inositol-1-monophosphatase, MAGNESIUM ION | | Authors: | Huang, Y.H, Loll, B, Wahl, M.C. | | Deposit date: | 2018-11-29 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.245 Å) | | Cite: | Structural basis for the function of SuhB as a transcription factor in ribosomal RNA synthesis.
Nucleic Acids Res., 47, 2019
|
|
5XGT
 
 | |
7RMR
 
 | | Crystal structure of [I11L]cycloviolacin O2 | | Descriptor: | D-[I11L]cycloviolacin O2, THIOCYANATE ION, [I11L]cycloviolacin O2 | | Authors: | Huang, Y.H, Du, Q, Craik, D.J. | | Deposit date: | 2021-07-28 | | Release date: | 2021-09-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Enabling Efficient Folding and High-Resolution Crystallographic Analysis of Bracelet Cyclotides.
Molecules, 26, 2021
|
|
7RMQ
 
 | | Crystal structure of cycloviolacin O2 | | Descriptor: | Cycloviolacin O2, D-[I11L]cycloviolacin O2, FORMIC ACID | | Authors: | Huang, Y.H, Du, Q. | | Deposit date: | 2021-07-28 | | Release date: | 2021-09-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Enabling Efficient Folding and High-Resolution Crystallographic Analysis of Bracelet Cyclotides.
Molecules, 26, 2021
|
|
7RMS
 
 | | Crystal structure of [I11G]cycloviolacin O2 | | Descriptor: | D-[I11L]cycloviolacin O2, THIOCYANATE ION, [I11L]cycloviolacin O2 | | Authors: | Huang, Y.H, Du, Q, Craik, D.J. | | Deposit date: | 2021-07-28 | | Release date: | 2021-09-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Enabling Efficient Folding and High-Resolution Crystallographic Analysis of Bracelet Cyclotides.
Molecules, 26, 2021
|
|
6TQN
 
 | | rrn anti-termination complex without S4 | | Descriptor: | 30S ribosomal protein S10, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Huang, Y.H, Wahl, M.C, Loll, B, Hilal, T, Said, N. | | Deposit date: | 2019-12-17 | | Release date: | 2020-08-05 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure-Based Mechanisms of a Molecular RNA Polymerase/Chaperone Machine Required for Ribosome Biosynthesis.
Mol.Cell, 79, 2020
|
|
6TQO
 
 | | rrn anti-termination complex | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S4, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Huang, Y.H, Wahl, M.C, Loll, B, Hilal, T, Said, N. | | Deposit date: | 2019-12-17 | | Release date: | 2020-08-05 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure-Based Mechanisms of a Molecular RNA Polymerase/Chaperone Machine Required for Ribosome Biosynthesis.
Mol.Cell, 79, 2020
|
|
4F5D
 
 | | ERIS/STING in complex with ligand | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MAGNESIUM ION, Transmembrane protein 173 | | Authors: | Huang, Y.H, Liu, X.Y, Su, X.D. | | Deposit date: | 2012-05-13 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structural basis for the sensing and binding of cyclic di-GMP by STING
Nat.Struct.Mol.Biol., 19, 2012
|
|
4F5E
 
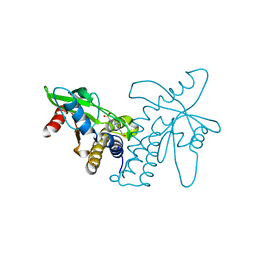 | | Crystal structure of ERIS/STING | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Transmembrane protein 173 | | Authors: | Huang, Y.H, Liu, X.Y, Su, X.D. | | Deposit date: | 2012-05-13 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | The structural basis for the sensing and binding of cyclic di-GMP by STING
Nat.Struct.Mol.Biol., 19, 2012
|
|
5YUO
 
 | |
6AEP
 
 | |
5DZL
 
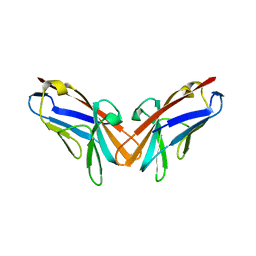 | | Crystal structure of the protein human CEACAM1 | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1 | | Authors: | Huang, Y.H, Russell, A, Gandhi, A.K, Kondo, Y, Chen, Q, Petsko, G.A, Blumberg, R.S. | | Deposit date: | 2015-09-25 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4006 Å) | | Cite: | CEACAM1 regulates TIM-3-mediated tolerance and exhaustion.
Nature, 517, 2015
|
|
4QXW
 
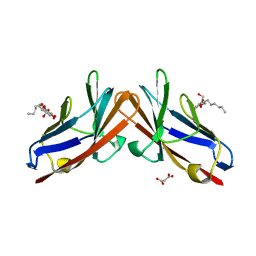 | | Crystal structure of the human CEACAM1 membrane distal amino terminal (N)-domain | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, MALONIC ACID, octyl beta-D-glucopyranoside | | Authors: | Huang, Y.H, Gandhi, A.K, Russell, A, Kondo, Y, Chen, Q, Petsko, G.A, Blumberg, R.S. | | Deposit date: | 2014-07-22 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | CEACAM1 regulates TIM-3-mediated tolerance and exhaustion.
Nature, 517, 2015
|
|
8WQ9
 
 | |
6IRQ
 
 | |
6KLK
 
 | |
6JDG
 
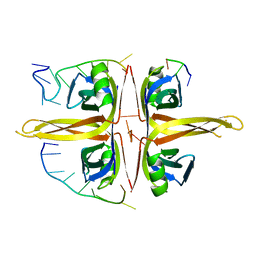 | | Complexed crystal structure of PaSSB with ssDNA dT20 at 2.39 angstrom resolution | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Single-stranded DNA-binding protein | | Authors: | Huang, Y.H, Huang, C.Y. | | Deposit date: | 2019-02-01 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.388 Å) | | Cite: | Complexed crystal structure of SSB reveals a novel single-stranded DNA binding mode (SSB)3:1: Phe60 is not crucial for defining binding paths.
Biochem.Biophys.Res.Commun., 520, 2019
|
|
6IB8
 
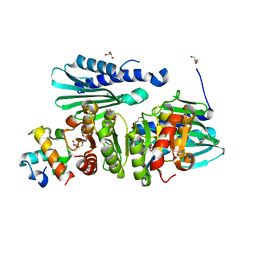 | | Structure of a complex of SuhB and NusA AR2 domain | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, GLYCEROL, Inositol-1-monophosphatase, ... | | Authors: | Huang, Y.H, Loll, B, Wahl, M.C. | | Deposit date: | 2018-11-29 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.646 Å) | | Cite: | Structural basis for the function of SuhB as a transcription factor in ribosomal RNA synthesis.
Nucleic Acids Res., 47, 2019
|
|
5YKD
 
 | |
5YNZ
 
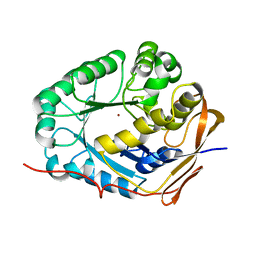 | | Crystal structure of the dihydroorotase domain (K1556A) of human CAD | | Descriptor: | CAD protein, ZINC ION | | Authors: | Huang, Y.H, Chen, K.L, Cheng, J.H, Huang, C.Y. | | Deposit date: | 2017-10-26 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.774 Å) | | Cite: | Crystal structures of monometallic dihydropyrimidinase and the human dihydroorotase domain K1556A mutant reveal no lysine carbamylation within the active site
Biochem. Biophys. Res. Commun., 505, 2018
|
|
6AEQ
 
 | |
6AJD
 
 | |
5YYU
 
 | |
7YM1
 
 | | Structure of SsbA protein in complex with the anticancer drug 5-fluorouracil | | Descriptor: | 5-FLUOROURACIL, GLYCEROL, Single-stranded DNA-binding protein | | Authors: | Huang, Y.H, Yang, P.C, Chiang, W.Y, Lin, E.S, Huang, C.Y. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal Structure of DNA Replication Protein SsbA Complexed with the Anticancer Drug 5-Fluorouracil.
Int J Mol Sci, 24, 2023
|
|
6XNT
 
 | | Crystal structure of I91A mutant of human CEACAM1 | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, octyl beta-D-glucopyranoside | | Authors: | Gandhi, A.K, Kim, W.M, Sun, Z.-Y, Huang, Y.H, Bonsor, D, Petsko, G.A, Kuchroo, V, Blumberg, R.S. | | Deposit date: | 2020-07-04 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of the dynamic human CEACAM1 monomer-dimer equilibrium.
Commun Biol, 4, 2021
|
|
