5JDK
 
 | |
3MGE
 
 | | X-ray Structure of Hexameric HIV-1 CA | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein p24 | | Authors: | Pornillos, O. | | Deposit date: | 2010-04-05 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Disulfide Bond Stabilization of the Hexameric Capsomer of Human Immunodeficiency Virus.
J.Mol.Biol., 401, 2010
|
|
5CD3
 
 | | Structure of immature VRC01-class antibody DRVIA7 | | Descriptor: | DRVIA7 Heavy Chain, DRVIA7 Light Chain | | Authors: | Kong, L, Wilson, I.A. | | Deposit date: | 2015-07-02 | | Release date: | 2016-04-06 | | Last modified: | 2018-09-05 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Key gp120 Glycans Pose Roadblocks to the Rapid Development of VRC01-Class Antibodies in an HIV-1-Infected Chinese Donor.
Immunity, 44, 2016
|
|
5CD5
 
 | |
3H47
 
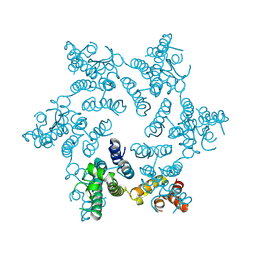 | | X-ray Structure of Hexameric HIV-1 CA | | Descriptor: | Capsid protein p24 | | Authors: | Pornillos, O. | | Deposit date: | 2009-04-18 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structures of the hexameric building block of the HIV capsid.
Cell(Cambridge,Mass.), 137, 2009
|
|
3GV2
 
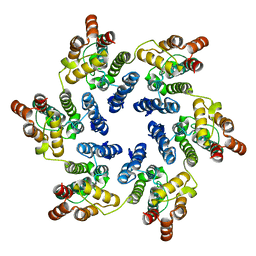 | | X-ray Structure of Hexameric HIV-1 CA | | Descriptor: | Capsid protein p24,Carbon dioxide-concentrating mechanism protein CcmK homolog 4 | | Authors: | Kelly, B.N. | | Deposit date: | 2009-03-30 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | X-ray structures of the hexameric building block of the HIV capsid.
Cell(Cambridge,Mass.), 137, 2009
|
|
7XHJ
 
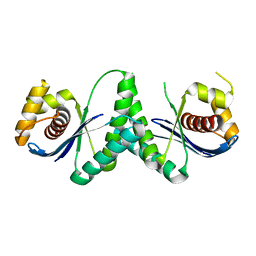 | | Crystal structure of RuvC from Deinococcus radiodurans | | Descriptor: | Crossover junction endodeoxyribonuclease RuvC | | Authors: | Qin, C, Zhao, Y. | | Deposit date: | 2022-04-08 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Characterization of the Holliday Junction Resolvase RuvC from Deinococcus radiodurans.
Microorganisms, 10, 2022
|
|
3H4E
 
 | | X-ray Structure of Hexameric HIV-1 CA | | Descriptor: | Capsid protein p24 | | Authors: | Pornillos, O. | | Deposit date: | 2009-04-19 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | X-ray structures of the hexameric building block of the HIV capsid.
Cell(Cambridge,Mass.), 137, 2009
|
|
7U2E
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody ADI-55688 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADI-55688 heavy chain, ADI-55688 light chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2022-02-23 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A broad and potent neutralization epitope in SARS-related coronaviruses.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7U2D
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody ADG20 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADG20 heavy chain, ADG20 light chain, ... | | Authors: | Zhu, X, Yuan, M, Wilson, I.A. | | Deposit date: | 2022-02-23 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | A broad and potent neutralization epitope in SARS-related coronaviruses.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8H6P
 
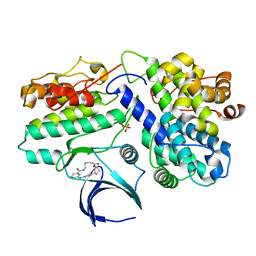 | | Complex structure of CDK2/Cyclin E1 and a potent, selective macrocyclic inhibitor | | Descriptor: | (7S,10R)-11-oxa-2,4,5,13,17,23-hexaazatetracyclo[17.3.1.1~3,6~.1~7,10~]pentacosa-1(23),3(25),5,19,21-pentaene-12,18-dione, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | Ren, X. | | Deposit date: | 2022-10-18 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Accelerated Discovery of Macrocyclic CDK2 Inhibitor QR-6401 by Generative Models and Structure-Based Drug Design.
Acs Med.Chem.Lett., 14, 2023
|
|
8H6T
 
 | | Complex structure of CDK2/Cyclin E1 and a potent, selective small molecule inhibitor | | Descriptor: | (1R,3S)-3-{3-[(pyridin-2-yl)amino]-1H-pyrazol-5-yl}cyclopentyl propan-2-ylcarbamate, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | Ren, X. | | Deposit date: | 2022-10-18 | | Release date: | 2023-02-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Accelerated Discovery of Macrocyclic CDK2 Inhibitor QR-6401 by Generative Models and Structure-Based Drug Design.
Acs Med.Chem.Lett., 14, 2023
|
|
5B7J
 
 | | Structure model of Sap1-DNA complex | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*TP*TP*GP*TP*TP*TP*TP*G)-3'), DNA (5'-D(*CP*AP*AP*AP*AP*CP*AP*AP*TP*AP*TP*T)-3'), Switch-activating protein 1 | | Authors: | Jin, C, Hu, Y, Ding, J, Zhang, Y. | | Deposit date: | 2016-06-07 | | Release date: | 2017-02-22 | | Last modified: | 2017-05-24 | | Method: | SOLUTION NMR | | Cite: | Sap1 is a replication-initiation factor essential for the assembly of pre-replicative complex in the fission yeast Schizosaccharomyces pombe
J. Biol. Chem., 292, 2017
|
|
4JM2
 
 | |
4JM4
 
 | | Crystal Structure of PGT 135 Fab | | Descriptor: | PGT 135 Heavy Chain, PGT 135 Light Chain | | Authors: | Kong, L, Wilson, I.A. | | Deposit date: | 2013-03-13 | | Release date: | 2013-05-29 | | Last modified: | 2013-07-17 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120.
Nat.Struct.Mol.Biol., 20, 2013
|
|
7COC
 
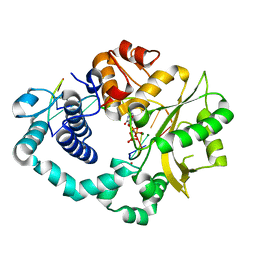 | | Ternary complex of DNA polymerase Mu (K438A/Q441A) with 1-nt gapped DNA (T:dGMPNPP) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*CP*GP*GP*CP*TP*TP*AP*CP*G)-3'), DNA (5'-D(*CP*GP*TP*A)-3'), ... | | Authors: | Guo, M, Zhao, Y. | | Deposit date: | 2020-08-04 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of genome instability mediated by human DNA polymerase mu misincorporation.
Nat Commun, 12, 2021
|
|
7COB
 
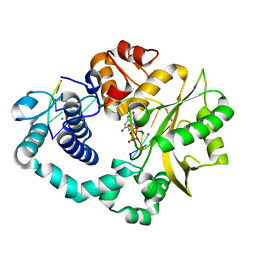 | | Ternary complex of DNA polymerase Mu (Q441A) with 1-nt gapped DNA (T:dGMPNPP) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*CP*GP*GP*CP*TP*TP*AP*CP*G)-3'), DNA (5'-D(*CP*GP*TP*A)-3'), ... | | Authors: | Guo, M, Zhao, Y. | | Deposit date: | 2020-08-04 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Mechanism of genome instability mediated by human DNA polymerase mu misincorporation.
Nat Commun, 12, 2021
|
|
7CO6
 
 | |
7CO9
 
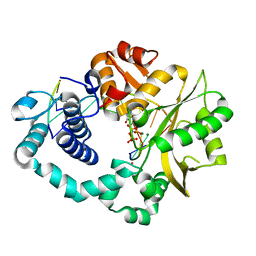 | | Ternary complex of DNA polymerase Mu with 1-nt gapped DNA (T:dGMPNPP) and Mg | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*CP*GP*GP*CP*TP*TP*AP*CP*G)-3'), DNA (5'-D(*CP*GP*TP*A)-3'), ... | | Authors: | Guo, M, Zhao, Y. | | Deposit date: | 2020-08-04 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Mechanism of genome instability mediated by human DNA polymerase mu misincorporation.
Nat Commun, 12, 2021
|
|
7CO8
 
 | | Ternary complex of DNA polymerase Mu with 2-nt gapped DNA (T:dGMPNPP) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*CP*GP*GP*CP*TP*TP*TP*AP*CP*G)-3'), DNA (5'-D(*CP*GP*TP*A)-3'), ... | | Authors: | Guo, M, Zhao, Y. | | Deposit date: | 2020-08-04 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Mechanism of genome instability mediated by human DNA polymerase mu misincorporation.
Nat Commun, 12, 2021
|
|
7COA
 
 | | Ternary complex of DNA polymerase Mu with 1-nt gapped DNA (T:dGMPNPP) and Mn | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*CP*GP*GP*CP*TP*TP*AP*CP*G)-3'), DNA (5'-D(*CP*GP*TP*A)-3'), ... | | Authors: | Guo, M, Zhao, Y. | | Deposit date: | 2020-08-04 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Mechanism of genome instability mediated by human DNA polymerase mu misincorporation.
Nat Commun, 12, 2021
|
|
7COD
 
 | |
6WO4
 
 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2 core from genotype 6a bound to broadly neutralizing antibody HC11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein E2, ... | | Authors: | Tzarum, N, Wilson, I.A, Law, M. | | Deposit date: | 2020-04-24 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | An alternate conformation of HCV E2 neutralizing face as an additional vaccine target.
Sci Adv, 6, 2020
|
|
6WO3
 
 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2 core from genotype 6a bound to broadly neutralizing antibody U1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein E2, ... | | Authors: | Tzarum, N, Wilson, I.A, Law, M. | | Deposit date: | 2020-04-24 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | An alternate conformation of HCV E2 neutralizing face as an additional vaccine target.
Sci Adv, 6, 2020
|
|
6WOQ
 
 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2 core from genotype 1a bound to neutralizing antibody HC1AM and non neutralizing antibody E1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein E2, ... | | Authors: | Tzarum, N, Wilson, I.A, Law, M. | | Deposit date: | 2020-04-25 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.667 Å) | | Cite: | An alternate conformation of HCV E2 neutralizing face as an additional vaccine target.
Sci Adv, 6, 2020
|
|
