8K9Q
 
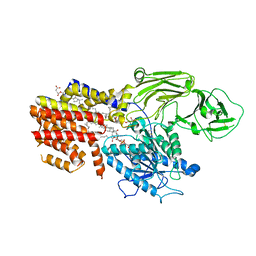 | | Cryo-EM structure of the GPI inositol-deacylase (PGAP1/Bst1) from Chaetomium thermophilum | | Descriptor: | (2~{S})-2-azanyl-3-[[(2~{R})-3-hexadecanoyloxy-2-[(~{Z})-octadec-9-enoyl]oxy-propoxy]-oxidanyl-phosphoryl]oxy-propanoic acid, CHOLESTEROL HEMISUCCINATE, GPI inositol-deacylase,fused thermostable green fluorescent protein | | Authors: | Hong, J, Li, T, Qu, Q, Li, D. | | Deposit date: | 2023-08-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Molecular basis of the inositol deacylase PGAP1 involved in quality control of GPI-AP biogenesis.
Nat Commun, 15, 2024
|
|
2JXN
 
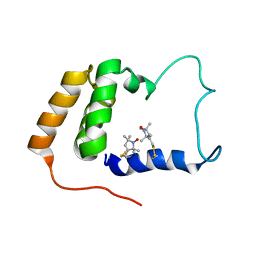 | | Solution Structure of S. cerevisiae PDCD5-like Protein Ymr074cp | | Descriptor: | S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, Uncharacterized protein YMR074C | | Authors: | Hong, J, Zhang, J, Liu, Z, Shi, Y, Wu, J. | | Deposit date: | 2007-11-23 | | Release date: | 2008-12-02 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Dynamics of S. cerevisiae PDCD5-like Protein Ymr074cp Determined by Heteronuclear NMR Spectroscopy
To be Published
|
|
2MLD
 
 | |
2MLA
 
 | |
2LY8
 
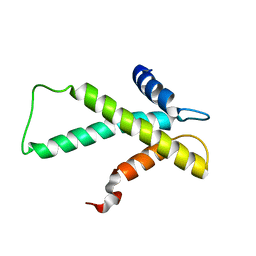 | | The budding yeast chaperone Scm3 recognizes the partially unfolded dimer of the centromere-specific Cse4/H4 histone variant | | Descriptor: | Budding yeast chaperone Scm3 | | Authors: | Hong, J, Feng, H, Zhou, Z, Ghirlando, R, Bai, Y. | | Deposit date: | 2012-09-13 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Identification of Functionally Conserved Regions in the Structure of the Chaperone/CenH3/H4 Complex.
J.Mol.Biol., 425, 2013
|
|
2MVA
 
 | |
2JOJ
 
 | |
2KCR
 
 | |
8GQM
 
 | |
8GQK
 
 | |
8GQI
 
 | |
8GQJ
 
 | |
8GQF
 
 | |
8GQH
 
 | |
8GQN
 
 | |
8GQL
 
 | |
8GQG
 
 | |
6J2V
 
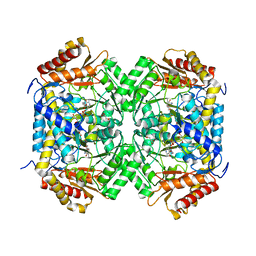 | | GABA aminotransferase from Corynebacterium glutamicum | | Descriptor: | 4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]BUTANOIC ACID, GLYCEROL, PLP-dependent aminotransferases | | Authors: | Hong, J, Kim, K.J. | | Deposit date: | 2019-01-03 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of gamma-aminobutyrate aminotransferase in complex with a PLP-GABA adduct from Corynebacterium glutamicum.
Biochem.Biophys.Res.Commun., 514, 2019
|
|
7CW4
 
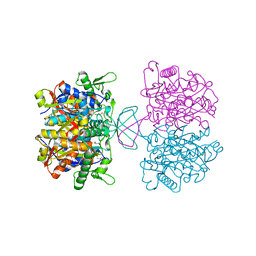 | | Acetyl-CoA acetyltransferase from Bacillus cereus ATCC 14579 | | Descriptor: | Acetyl-CoA acetyltransferase, GLYCEROL | | Authors: | Hong, J, Kim, K.J. | | Deposit date: | 2020-08-27 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of an acetyl-CoA acetyltransferase from PHB producing bacterium Bacillus cereus ATCC 14579.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
7CW5
 
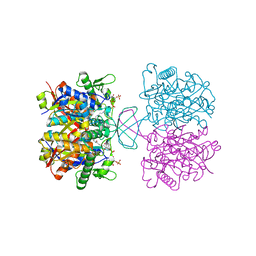 | | Acetyl-CoA acetyltransferase from Bacillus cereus ATCC 14579 | | Descriptor: | Acetyl-CoA acetyltransferase, COENZYME A | | Authors: | Hong, J, Kim, K.J. | | Deposit date: | 2020-08-27 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an acetyl-CoA acetyltransferase from PHB producing bacterium Bacillus cereus ATCC 14579.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
8TUM
 
 | |
8TUX
 
 | |
8TUW
 
 | |
7WZ6
 
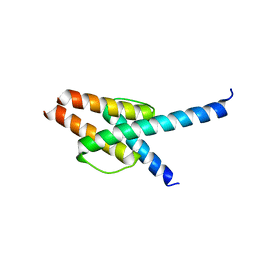 | | Crystal structure of MyoD-E47 | | Descriptor: | Isoform E47 of Transcription factor E2-alpha, Myoblast determination protein 1 | | Authors: | Zhong, J, Huang, Y, Ma, J. | | Deposit date: | 2022-02-17 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of the bHLH domains of MyoD-E47 heterodimer.
Biochem.Biophys.Res.Commun., 621, 2022
|
|
8K9R
 
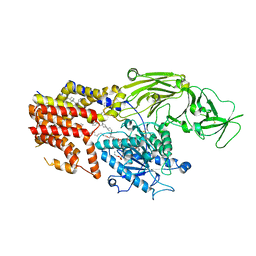 | | Cryo EM structure of the products-bound PGAP1(Bst1)-H443N from Chaetomium thermophilum | | Descriptor: | 2-amino-2-deoxy-alpha-D-glucopyranose, 2-azanylethyl [(2R,3S,4S,5S,6S)-3,4,5,6-tetrakis(oxidanyl)oxan-2-yl]methyl hydrogen phosphate, 2-azanylethyl [(2~{S},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-2,4,5-tris(oxidanyl)oxan-3-yl] hydrogen phosphate, ... | | Authors: | Li, T, Hong, J, Qu, Q, Li, D. | | Deposit date: | 2023-08-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Molecular basis of the inositol deacylase PGAP1 involved in quality control of GPI-AP biogenesis.
Nat Commun, 15, 2024
|
|
