1TUK
 
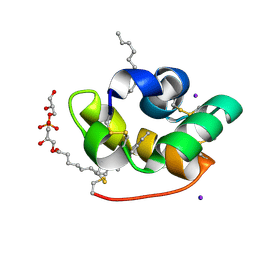 | | Crystal structure of liganded type 2 non specific lipid transfer protein from wheat | | Descriptor: | 1-MYRISTOYL-2-HYDROXY-SN-GLYCERO-3-[PHOSPHO-RAC-(1-GLYCEROL)], IODIDE ION, Nonspecific lipid-transfer protein 2G | | Authors: | Hoh, F, Pons, J.L, Gautier, M.F, De Lamotte, F, Dumas, C. | | Deposit date: | 2004-06-25 | | Release date: | 2005-04-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structure of a liganded type 2 non-specific lipid-transfer protein from wheat and the molecular basis of lipid binding.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
6R5J
 
 | |
1JSG
 
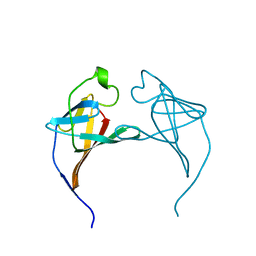 | | CRYSTAL STRUCTURE OF P14TCL1, AN ONCOGENE PRODUCT INVOLVED IN T-CELL PROLYMPHOCYTIC LEUKEMIA, REVEALS A NOVEL B-BARREL TOPOLOGY | | Descriptor: | ONCOGENE PRODUCT P14TCL1 | | Authors: | Hoh, F, Yang, Y.-S, Guignard, L, Padilla, A, Stern, R.-H, Lhoste, J.-M, Van Tilbeurgh, H. | | Deposit date: | 1997-12-03 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of p14TCL1, an oncogene product involved in T-cell prolymphocytic leukemia, reveals a novel beta-barrel topology.
Structure, 6, 1998
|
|
3F45
 
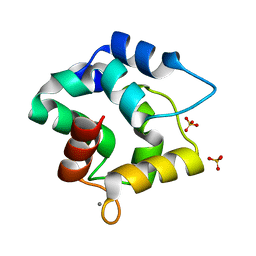 | | Structure of the R75A mutant of rat alpha-Parvalbumin | | Descriptor: | CALCIUM ION, Parvalbumin alpha, SULFATE ION | | Authors: | Hoh, F, Padilla, A. | | Deposit date: | 2008-10-31 | | Release date: | 2009-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Removing the invariant salt bridge of parvalbumin increases flexibility in the AB-loop structure
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3F6N
 
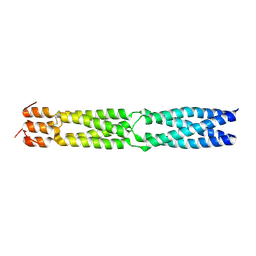 | |
1T7H
 
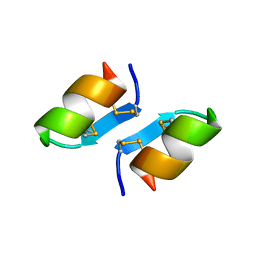 | | X-ray structure of [Lys(-2)-Arg(-1)-des(17-21)]-endothelin-1 peptide | | Descriptor: | Endothelin-1 | | Authors: | Hoh, F, Cerdan, R, Kaas, Q, Nishi, Y, Chiche, L, Kubo, S, Chino, N, Kobayashi, Y, Dumas, C, Aumelas, A. | | Deposit date: | 2004-05-10 | | Release date: | 2004-12-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | High-resolution X-ray structure of the unexpectedly stable dimer of the [Lys(-2)-Arg(-1)-des(17-21)]endothelin-1 peptide
Biochemistry, 43, 2004
|
|
8ACX
 
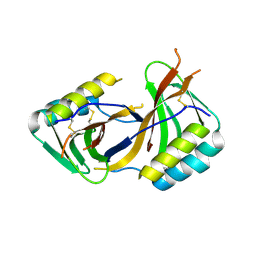 | | Pathogen effector of Zymoseptoria tritici: Zt-KP4 | | Descriptor: | Hce2 domain-containing protein | | Authors: | Hoh, F, Padilla, A, De Guillen, K. | | Deposit date: | 2022-07-07 | | Release date: | 2024-01-17 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Zymoseptoria tritici effectors structurally related to killer proteins UmV-KP4 and UmV-KP6 are toxic to fungi, and define extended protein families in fungi
Biorxiv, 2024
|
|
7ZVM
 
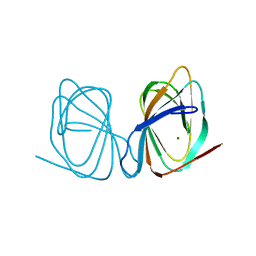 | |
7ZVY
 
 | | Thermococcus kadokarensis phosphomannose isomerase | | Descriptor: | Cupin_2 domain-containing protein, ZINC ION | | Authors: | Hoh, f, Calio, A. | | Deposit date: | 2022-05-17 | | Release date: | 2022-08-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Unravelling the Adaptation Mechanisms to High Pressure in Proteins.
Int J Mol Sci, 23, 2022
|
|
3K4T
 
 | |
4JMO
 
 | |
4JWL
 
 | |
4JXH
 
 | |
4JMI
 
 | |
4L5M
 
 | |
2GFI
 
 | | Crystal structure of the phytase from D. castellii at 2.3 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, phytase | | Authors: | Hoh, F. | | Deposit date: | 2006-03-22 | | Release date: | 2007-03-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure of Debaryomyces castellii CBS 2923 phytase.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
9Q8J
 
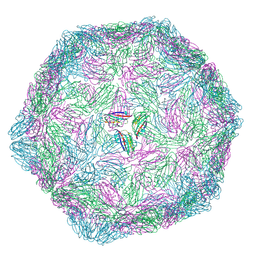 | | CryoEM structure of modified Turnip Yellows Virus devoid of minor capsid protein readthrough domain | | Descriptor: | Minor capsid readthrough protein | | Authors: | Trapani, S, Lai Kee Him, J, Hoh, F, Brault, V, Bron, P. | | Deposit date: | 2025-02-24 | | Release date: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structure of the turnip yellows virus particles.
Virology, 607, 2025
|
|
5LXA
 
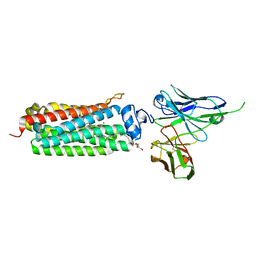 | | CRYSTAL STRUCTURE OF HUMAN ADIPONECTIN RECEPTOR 2 IN COMPLEX WITH A C18 FREE FATTY ACID AT 3.0 ANGSTROM RESOLUTION | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adiponectin receptor protein 2, Anti-CD30 moab Ki-4 scFv, ... | | Authors: | Vasiliauskaite-Brooks, I, Leyrat, C, Hoh, F, Granier, S. | | Deposit date: | 2016-09-20 | | Release date: | 2017-03-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into adiponectin receptors suggest ceramidase activity.
Nature, 6, 2017
|
|
6TY0
 
 | |
6TY2
 
 | |
8AA8
 
 | |
8A8S
 
 | |
7P8X
 
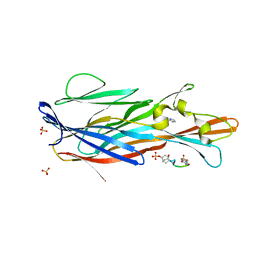 | | Crystal Structure of leukotoxin LukE from Staphylococcus aureus in complex with a doubly sulfated CCR2 N-terminal peptide | | Descriptor: | C-C chemokine receptor type 2, IMIDAZOLE, Leucotoxin LukEv, ... | | Authors: | Lambey, P, Hoh, F, Peysson, F, Granier, S, Leyrat, C. | | Deposit date: | 2021-07-23 | | Release date: | 2022-04-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights into recognition of chemokine receptors by Staphylococcus aureus leukotoxins.
Elife, 11, 2022
|
|
7P8U
 
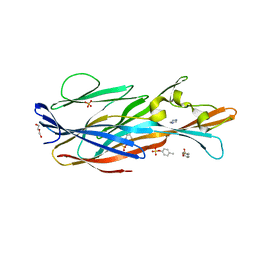 | | Crystal Structure of leukotoxin LukE from Staphylococcus aureus in complex with p-cresyl sulfate | | Descriptor: | (4-methylphenyl) hydrogen sulfate, DI(HYDROXYETHYL)ETHER, IMIDAZOLE, ... | | Authors: | Lambey, P, Hoh, F, Peysson, F, Granier, S, Leyrat, C. | | Deposit date: | 2021-07-23 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into recognition of chemokine receptors by Staphylococcus aureus leukotoxins.
Elife, 11, 2022
|
|
7P93
 
 | | Crystal Structure of leukotoxin LukE from Staphylococcus aureus in complex with a sulfated ACKR1 N-terminal peptide | | Descriptor: | Atypical chemokine receptor 1, Leucotoxin LukEv | | Authors: | Lambey, P, Hoh, F, Peysson, F, Granier, S, Leyrat, C. | | Deposit date: | 2021-07-23 | | Release date: | 2022-04-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights into recognition of chemokine receptors by Staphylococcus aureus leukotoxins.
Elife, 11, 2022
|
|
