7U2R
 
 | | Structure of Paenibacillus sp. J14 Apyc1 | | Descriptor: | Apyc1, ZINC ION | | Authors: | Hobbs, S.J, Wein, T, Lu, A, Morehouse, B.R, Schnabel, J, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2022-02-24 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Phage anti-CBASS and anti-Pycsar nucleases subvert bacterial immunity.
Nature, 605, 2022
|
|
7U2S
 
 | | Structure of Paenibacillus xerothermodurans Apyc1 in the apo state | | Descriptor: | Apyc1, ZINC ION | | Authors: | Hobbs, S.J, Wein, T, Lu, A, Morehouse, B.R, Schnabel, J, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2022-02-24 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Phage anti-CBASS and anti-Pycsar nucleases subvert bacterial immunity.
Nature, 605, 2022
|
|
9BKQ
 
 | | Structure of penguinpox cGAMP PDE in apo and post reaction states | | Descriptor: | 9-{3-O-[(S)-thiophosphono]-alpha-L-lyxofuranosyl}-9H-purin-6-amine, GUANINE, Penguinpox cGAMP PDE | | Authors: | Hobbs, S.J, Nomburg, J, Doudna, J.A, Kranzusch, P.J. | | Deposit date: | 2024-04-29 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Animal and bacterial viruses share conserved mechanisms of immune evasion.
Cell, 187, 2024
|
|
9CIW
 
 | | Penguinpox cGAMP PDE H72A mutant in complex with 2'3'-cGAMP | | Descriptor: | Penguinpox cGAMP PDE, [(2~{R},3~{R},4~{R},5~{S})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl [(2~{R},3~{R},4~{S},5~{R})-2-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-5-(hydroxymethyl)-4-oxidanyl-oxolan-3-yl] hydrogen phosphate | | Authors: | Hobbs, S.J, Nomburg, J, Doudna, J.A, Kranzusch, P.J. | | Deposit date: | 2024-07-05 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Animal and bacterial viruses share conserved mechanisms of immune evasion.
Cell, 187, 2024
|
|
7T26
 
 | | Structure of phage FBB1 anti-CBASS nuclease Acb1 in apo state | | Descriptor: | Acb1 | | Authors: | Hobbs, S.J, Wein, T, Lu, A, Morehouse, B.R, Schnabel, J, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-04-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Phage anti-CBASS and anti-Pycsar nucleases subvert bacterial immunity.
Nature, 605, 2022
|
|
7T27
 
 | | Structure of phage FBB1 anti-CBASS nuclease Acb1-3'3'-cGAMP complex in post reaction state | | Descriptor: | Acb1, SULFATE ION, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-2-[[[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-2-(hydroxymethyl)-4-oxidanyl-oxolan-3-yl]oxy-sulfanyl-phosphoryl]oxymethyl]-4-oxidanyl-oxolan-3-yl]oxy-sulfanyl-phosphinic acid | | Authors: | Hobbs, S.J, Wein, T, Lu, A, Morehouse, B.R, Schnabel, J, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-04-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Phage anti-CBASS and anti-Pycsar nucleases subvert bacterial immunity.
Nature, 605, 2022
|
|
7T28
 
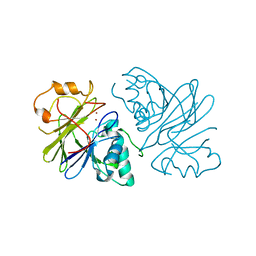 | | Structure of phage Bsp38 anti-Pycsar nuclease Apyc1 in apo state | | Descriptor: | Putative metal-dependent hydrolase, ZINC ION | | Authors: | Hobbs, S.J, Wein, T, Lu, A, Morehouse, B.R, Schnabel, J, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Phage anti-CBASS and anti-Pycsar nucleases subvert bacterial immunity.
Nature, 605, 2022
|
|
9B7D
 
 | | Structure of ThsB-Tad3 complex | | Descriptor: | Putative cyclic ADP-D-ribose synthase ThsB1, Tad3 | | Authors: | Hobbs, S.J, Tan, J.M.J, Yirmiya, E, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2024-03-27 | | Release date: | 2025-01-22 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-guided discovery of viral proteins that inhibit host immunity.
Cell, 188, 2025
|
|
9NYI
 
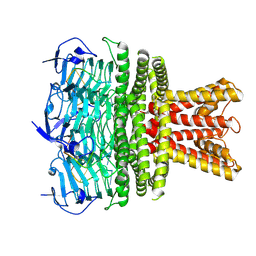 | | Structure of HalA in complex with oligodeoxyadenylate | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*A)-3'), Structure of HalA in complex with oligodeoxyadenylate | | Authors: | Tan, J.M.J, Melamed, S, Cofsky, J.C, Syangtan, D, Hobbs, S.J, Del Marmol, J, Jost, M, Kruse, A.C, Sorek, S, Kranzusch, P.J. | | Deposit date: | 2025-03-27 | | Release date: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (1.98 Å) | | Cite: | Structure of Hailong HalA in complex with oligodeoxyadenylate
To Be Published
|
|
9E4W
 
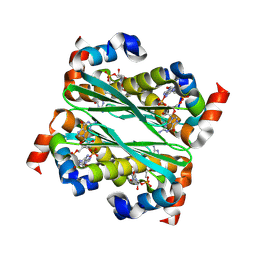 | | Structure of Bacillus phage SPO1 anti-CBASS 4 (Acb4) in complex with 3'3'-cGAMP | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, anti-CBASS 4 (Acb4) | | Authors: | Chang, R.B, Toyoda, H.C, Hobbs, S.J, Richmond-Buccola, D, Wein, T, Burger, N, Chouchani, E.T, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2024-10-25 | | Release date: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A widespread family of viral sponge proteins reveals specific inhibition of nucleotide signals in anti-phage defense.
Biorxiv, 2024
|
|
9DBJ
 
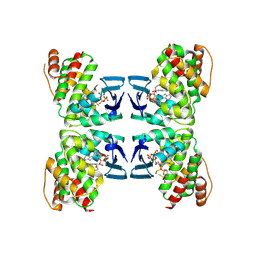 | | Structure of Hailong HalB R164A mutant with non-hydrolyzable dATP | | Descriptor: | (2R)-2-amino-3-(4-{[(R)-{[(2R,3R,5R)-5-(6-amino-9H-purin-9-yl)-3-hydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl]oxy}phenyl)propanoic acid (non-preferred name), (2R)-3-(4-{[(S)-{[(2R,3R,5R)-5-(6-amino-9H-purin-9-yl)-3-hydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl]oxy}phenyl)-2-{[(2R)-pyrrolidine-2-carbonyl]amino}propanoic acid (non-preferred name), 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Tan, J.M.J, Melamed, S, Cofsky, J.C, Hobbs, S.J, Kruse, A.C, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2024-08-23 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A DNA-gated molecular guard controls bacterial Hailong anti-phage defence.
Nature, 2025
|
|
9DBI
 
 | | Structure of Hailong HalB D21A/D23A mutant | | Descriptor: | HalB, PHOSPHATE ION | | Authors: | Tan, J.M.J, Melamed, S, Cofsky, J.C, Hobbs, S.J, Kruse, A.C, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2024-08-23 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A DNA-gated molecular guard controls bacterial Hailong anti-phage defence.
Nature, 2025
|
|
9DBH
 
 | | Structure of Hailong HalB in complex with oligodeoxyadenylate | | Descriptor: | DNA (5'-D(P*AP*AP*AP*AP*AP*A)-3'), HalB, MANGANESE (II) ION | | Authors: | Tan, J.M.J, Melamed, S, Cofsky, J.C, Hobbs, S.J, Kruse, A.C, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2024-08-23 | | Release date: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure of HalB in complex with oligodeoxyadenylate
To Be Published
|
|
7UAV
 
 | | Structure of Clostridium botulinum prophage Tad1 in apo state | | Descriptor: | ABC transporter ATPase | | Authors: | Lu, A, Leavitt, A, Yirmiya, E, Amitai, G, Garb, J, Morehouse, B.R, Hobbs, S.J, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2022-03-14 | | Release date: | 2022-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Viruses inhibit TIR gcADPR signalling to overcome bacterial defence.
Nature, 611, 2022
|
|
7UAW
 
 | | Structure of Clostridium botulinum prophage Tad1 in complex with 1''-2' gcADPR | | Descriptor: | (1S,3R,4R,6R,9S,11R,14R,15S,16R,18R)-4-(6-amino-9H-purin-9-yl)-9,11,15,16,18-pentahydroxy-2,5,8,10,12,17-hexaoxa-9lambda~5~,11lambda~5~-diphosphatricyclo[12.2.1.1~3,6~]octadecane-9,11-dione, ABC transporter ATPase | | Authors: | Lu, A, Leavitt, A, Yirmiya, E, Amitai, G, Garb, J, Morehouse, B.R, Hobbs, S.J, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2022-03-14 | | Release date: | 2022-10-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Viruses inhibit TIR gcADPR signalling to overcome bacterial defence.
Nature, 611, 2022
|
|
8SMF
 
 | | Structure of SPO1 phage Tad2 in complex with 1''-3' gcADPR | | Descriptor: | (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione, Gp34.65, MAGNESIUM ION | | Authors: | Lu, A, Yirmiya, E, Leavitt, A, Avraham, C, Osterman, I, Garb, J, Antine, S.P, Mooney, S.E, Hobbs, S.J, Amitai, G, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2023-04-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Phages overcome bacterial immunity via diverse anti-defence proteins.
Nature, 625, 2024
|
|
8SMG
 
 | | Structure of SPO1 phage Tad2 in complex with 1''-2' gcADPR | | Descriptor: | (1S,3R,4R,6R,9S,11R,14R,15S,16R,18R)-4-(6-amino-9H-purin-9-yl)-9,11,15,16,18-pentahydroxy-2,5,8,10,12,17-hexaoxa-9lambda~5~,11lambda~5~-diphosphatricyclo[12.2.1.1~3,6~]octadecane-9,11-dione, Gp34.65 | | Authors: | Lu, A, Yirmiya, E, Leavitt, A, Avraham, C, Osterman, I, Garb, J, Antine, S.P, Mooney, S.E, Hobbs, S.J, Amitai, G, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2023-04-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phages overcome bacterial immunity via diverse anti-defence proteins.
Nature, 625, 2024
|
|
8SMD
 
 | | Structure of Clostridium botulinum prophage Tad1 in complex with 1''-3' gcADPR | | Descriptor: | (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione, ABC transporter ATPase | | Authors: | Lu, A, Yirmiya, E, Leavitt, A, Avraham, C, Osterman, I, Garb, J, Antine, S.P, Mooney, S.E, Hobbs, S.J, Amitai, G, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2023-04-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phages overcome bacterial immunity via diverse anti-defence proteins.
Nature, 625, 2024
|
|
8SME
 
 | | Structure of SPO1 phage Tad2 in apo state | | Descriptor: | Gp34.65 | | Authors: | Lu, A, Yirmiya, E, Leavitt, A, Avraham, C, Osterman, I, Garb, J, Antine, S.P, Mooney, S.E, Hobbs, S.J, Amitai, G, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2023-04-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Phages overcome bacterial immunity via diverse anti-defence proteins.
Nature, 625, 2024
|
|
8T9N
 
 | | Bacillus subtilis RsgI GGG mutant | | Descriptor: | Anti-sigma-I factor RsgI | | Authors: | Brogan, A.P, Habib, C, Hobbs, S.J, Kranzusch, P.J, Rudner, D.Z. | | Deposit date: | 2023-06-24 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bacterial SEAL domains undergo autoproteolysis and function in regulated intramembrane proteolysis.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
9ARD
 
 | |
8TTO
 
 | |
8U7I
 
 | | Structure of the phage immune evasion protein Gad1 bound to the Gabija GajAB complex | | Descriptor: | Endonuclease GajA, Gabija Anti-Defense 1, Gabija protein GajB | | Authors: | Antine, S.P, Johnson, A.G, Mooney, S.E, Mayer, M.L, Kranzusch, P.J. | | Deposit date: | 2023-09-15 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural basis of Gabija anti-phage defence and viral immune evasion.
Nature, 625, 2024
|
|
