6JLN
 
 | | XFEL structure of cyanobacterial photosystem II (1F state, dataset2) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
6JLJ
 
 | | XFEL structure of cyanobacterial photosystem II (dark state, dataset1) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
6JLM
 
 | | XFEL structure of cyanobacterial photosystem II (dark state, dataset2) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
6JLK
 
 | | XFEL structure of cyanobacterial photosystem II (1F state, dataset1) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
6JLO
 
 | | XFEL structure of cyanobacterial photosystem II (2F state, dataset2) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
6JLL
 
 | | XFEL structure of cyanobacterial photosystem II (2F state, dataset1) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
5Y50
 
 | | Crystal structure of eukaryotic MATE transporter AtDTX14 | | Descriptor: | Protein DETOXIFICATION 14 | | Authors: | Miyauchi, H, Kusakizako, T, Nishizawa, T, Ishitani, R, Nureki, O. | | Deposit date: | 2017-08-06 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for xenobiotic extrusion by eukaryotic MATE transporter
Nat Commun, 8, 2017
|
|
5X8W
 
 | | Crystal Structure of the mutant Human ROR gamma Ligand Binding Domain. | | Descriptor: | Nuclear receptor ROR-gamma, Nuclear receptor coactivator 1 | | Authors: | Noguchi, M, Nomura, A, Murase, K, Doi, S, Yamaguchi, K, Adachi, T. | | Deposit date: | 2017-03-03 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ternary complex of human ROR gamma ligand-binding domain, inverse agonist and SMRT peptide shows a unique mechanism of corepressor recruitment
Genes Cells, 22, 2017
|
|
5X8X
 
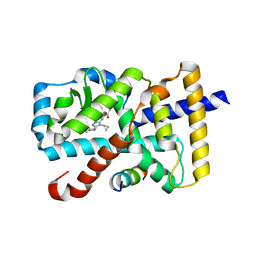 | | Crystal Structure of the mutant Human ROR gamma Ligand Binding Domain With Compound A. | | Descriptor: | (3R,4R)-4-[4-cyclopropyl-5-[3-(2-methylpropyl)cyclobutyl]-1,2,4-triazol-3-yl]-N-(2,4-dimethylphenyl)-1-ethanoyl-pyrrolidine-3-carboxamide, Nuclear receptor ROR-gamma, Nuclear receptor corepressor 2 | | Authors: | Noguchi, M, Nomura, A, Murase, K, Doi, S, Yamaguchi, K, Adachi, T. | | Deposit date: | 2017-03-03 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Ternary complex of human ROR gamma ligand-binding domain, inverse agonist and SMRT peptide shows a unique mechanism of corepressor recruitment
Genes Cells, 22, 2017
|
|
5B2G
 
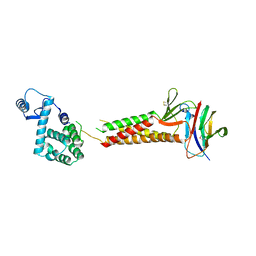 | | Crystal structure of human claudin-4 in complex with C-terminal fragment of Clostridium perfringens enterotoxin | | Descriptor: | Endolysin,Claudin-4, Heat-labile enterotoxin B chain | | Authors: | Shinoda, T, Kimura-Someya, T, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2016-01-15 | | Release date: | 2016-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for disruption of claudin assembly in tight junctions by an enterotoxin
Sci Rep, 6, 2016
|
|
5X8S
 
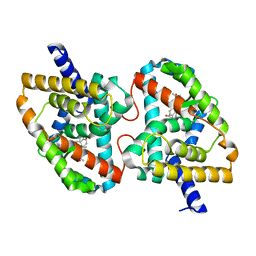 | | Crystal Structure of the mutant Human ROR gamma Ligand Binding Domain With Ursolic acid. | | Descriptor: | Nuclear receptor ROR-gamma, Ursolic acid | | Authors: | Noguchi, M, Nomura, A, Murase, K, Doi, S, Yamaguchi, K, Adachi, T. | | Deposit date: | 2017-03-03 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ternary complex of human ROR gamma ligand-binding domain, inverse agonist and SMRT peptide shows a unique mechanism of corepressor recruitment
Genes Cells, 22, 2017
|
|
5GXB
 
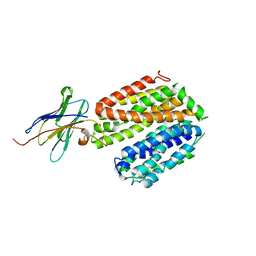 | | crystal structure of a LacY/Nanobody complex | | Descriptor: | Lactose permease, nanobody | | Authors: | Jiang, X, Wu, J.P, Yan, N, Kaback, H.R. | | Deposit date: | 2016-09-16 | | Release date: | 2016-10-26 | | Last modified: | 2022-10-12 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of a LacY-nanobody complex in a periplasmic-open conformation.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6KX8
 
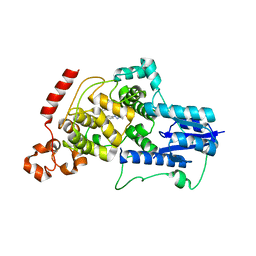 | | Crystal structure of mouse Cryptochrome 2 in complex with TH301 compound | | Descriptor: | 1-(4-chlorophenyl)-N-[2-(4-methoxyphenyl)-5,5-bis(oxidanylidene)-4,6-dihydrothieno[3,4-c]pyrazol-3-yl]cyclopentane-1-carboxamide, Cryptochrome-2 | | Authors: | Miller, S.A, Aikawa, Y, Hirota, T. | | Deposit date: | 2019-09-10 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Isoform-selective regulation of mammalian cryptochromes.
Nat.Chem.Biol., 16, 2020
|
|
6KX7
 
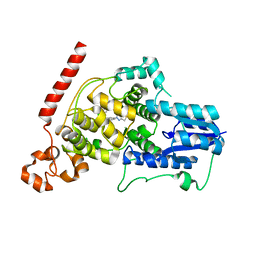 | | Crystal structure of mouse Cryptochrome 1 in complex with TH301 compound | | Descriptor: | 1-(4-chlorophenyl)-N-[2-(4-methoxyphenyl)-5,5-bis(oxidanylidene)-4,6-dihydrothieno[3,4-c]pyrazol-3-yl]cyclopentane-1-carboxamide, Cryptochrome-1 | | Authors: | Miller, S.A, Aikawa, Y, Hirota, T. | | Deposit date: | 2019-09-10 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Isoform-selective regulation of mammalian cryptochromes.
Nat.Chem.Biol., 16, 2020
|
|
6KX5
 
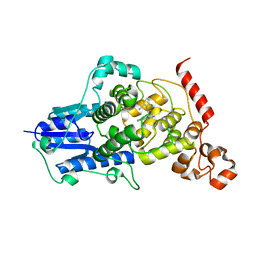 | |
6KX4
 
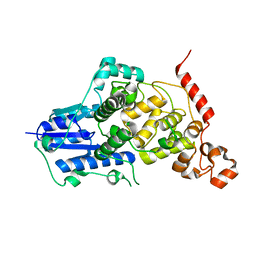 | |
6KX6
 
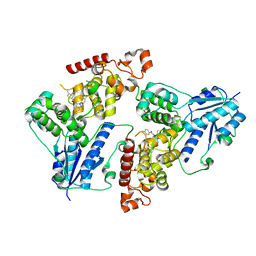 | | Crystal structure of mouse Cryptochrome 1 in complex with KL101 compound | | Descriptor: | Cryptochrome-1, ~{N}-[2-(2,4-dimethylphenyl)-4,6-dihydrothieno[3,4-c]pyrazol-3-yl]-3,4-dimethyl-benzamide | | Authors: | Miller, S.A, Aikawa, Y, Hirota, T. | | Deposit date: | 2019-09-10 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Isoform-selective regulation of mammalian cryptochromes.
Nat.Chem.Biol., 16, 2020
|
|
6A22
 
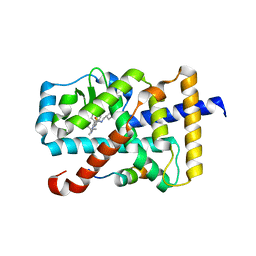 | | Ternary complex of Human ROR gamma Ligand Binding Domain With Compound T. | | Descriptor: | 2-[2-[1-~{tert}-butyl-5-(4-methoxyphenyl)pyrazol-4-yl]-1,3-thiazol-4-yl]-~{N}-(oxan-4-ylmethyl)ethanamide, Nuclear receptor ROR-gamma, Nuclear receptor corepressor 2 | | Authors: | Noguchi, M, Nomura, A, Doi, S, Yamaguchi, K, Adachi, T. | | Deposit date: | 2018-06-08 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Ternary crystal structure of human ROR gamma ligand-binding-domain, an inhibitor and corepressor peptide provides a new insight into corepressor interaction
Sci Rep, 8, 2018
|
|
6AK3
 
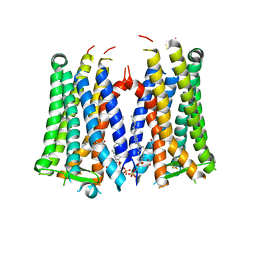 | | Crystal structure of the human prostaglandin E receptor EP3 bound to prostaglandin E2 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Prostaglandin E2 receptor EP3 subtype,Soluble cytochrome b562 | | Authors: | Morimoto, K, Suno, R, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-08-29 | | Release date: | 2018-12-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the endogenous agonist-bound prostanoid receptor EP3.
Nat. Chem. Biol., 15, 2019
|
|
5B3V
 
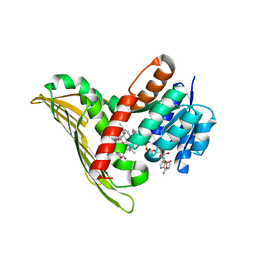 | |
5B3T
 
 | |
5B3U
 
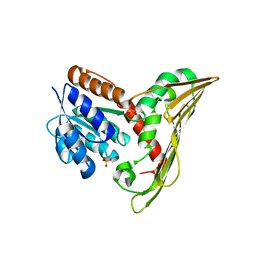 | |
5XSZ
 
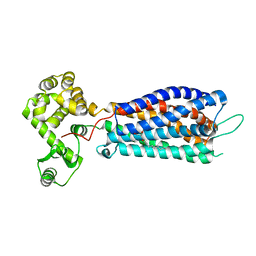 | | Crystal structure of zebrafish lysophosphatidic acid receptor LPA6 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Lysophosphatidic acid receptor 6a,Endolysin,Lysophosphatidic acid receptor 6a | | Authors: | Taniguchi, R, Nishizawa, T, Ishitani, R, Nureki, O. | | Deposit date: | 2017-06-16 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into ligand recognition by the lysophosphatidic acid receptor LPA6
Nature, 548, 2017
|
|
5ZLG
 
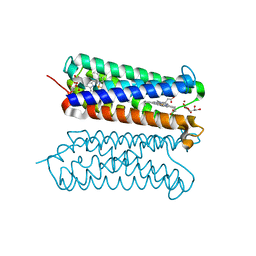 | | Human duodenal cytochrome b (Dcytb) in zinc ion and ascorbate bound form | | Descriptor: | ASCORBIC ACID, Cytochrome b reductase 1, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Ganasen, M, Togashi, H, Mauk, G.A, Shiro, Y, Sawai, H, Sugimoto, H. | | Deposit date: | 2018-03-27 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for promotion of duodenal iron absorption by enteric ferric reductase with ascorbate.
Commun Biol, 1, 2018
|
|
5ZLE
 
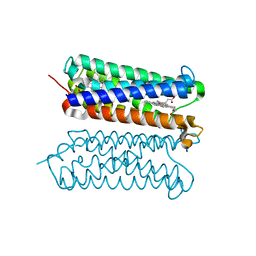 | | Human duodenal cytochrome b (Dcytb) in substrate free form | | Descriptor: | Cytochrome b reductase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ganasen, M, Togashi, H, Mauk, G.A, Shiro, Y, Sawai, H, Sugimoto, H. | | Deposit date: | 2018-03-27 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for promotion of duodenal iron absorption by enteric ferric reductase with ascorbate.
Commun Biol, 1, 2018
|
|
