2P8R
 
 | | Crystal structure of the C-terminal domain of C. elegans pre-mRNA splicing factor Prp8 carrying R2303K mutant | | Descriptor: | Pre-mRNA-splicing factor Prp8 | | Authors: | Zhang, L, Shen, J, Guarnieri, M.T, Heroux, A, Yang, K, Zhao, R. | | Deposit date: | 2007-03-22 | | Release date: | 2007-05-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the C-terminal domain of splicing factor Prp8 carrying retinitis pigmentosa mutants
Protein Sci., 16, 2007
|
|
2P87
 
 | | Crystal structure of the C-terminal domain of C. elegans pre-mRNA splicing factor Prp8 | | Descriptor: | Pre-mRNA-splicing factor Prp8 | | Authors: | Zhang, L, Shen, J, Guarnieri, M.T, Heroux, A, Yang, K, Zhao, R. | | Deposit date: | 2007-03-21 | | Release date: | 2007-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the C-terminal domain of splicing factor Prp8 carrying retinitis pigmentosa mutants
Protein Sci., 16, 2007
|
|
2R0Y
 
 | | Structure of the Rsc4 tandem bromodomain in complex with an acetylated H3 peptide | | Descriptor: | Chromatin structure-remodeling complex protein RSC4, Histone H3 peptide | | Authors: | VanDemark, A.P, Kasten, M.M, Ferris, E, Heroux, A, Hill, C.P, Cairns, B.R. | | Deposit date: | 2007-08-21 | | Release date: | 2007-10-30 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation.
Mol.Cell, 27, 2007
|
|
2R0V
 
 | | Structure of the Rsc4 tandem bromodomain acetylated at K25 | | Descriptor: | Chromatin structure-remodeling complex protein RSC4, SULFATE ION | | Authors: | VanDemark, A.P, Kasten, M.M, Ferris, E, Heroux, A, Hill, C.P, Cairns, B.R. | | Deposit date: | 2007-08-21 | | Release date: | 2007-10-30 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation.
Mol.Cell, 27, 2007
|
|
2R10
 
 | | Structure of an acetylated Rsc4 tandem bromodomain Histone Chimera | | Descriptor: | 1,2-ETHANEDIOL, Chromatin structure-remodeling complex protein RSC4, LINKER, ... | | Authors: | VanDemark, A.P, Kasten, M.M, Ferris, E, Heroux, A, Hill, C.P, Cairns, B.R. | | Deposit date: | 2007-08-21 | | Release date: | 2007-10-30 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation.
Mol.Cell, 27, 2007
|
|
2R0S
 
 | | Crystal Structure of the Rsc4 tandem bromodomain | | Descriptor: | Chromatin structure-remodeling complex protein RSC4 | | Authors: | VanDemark, A.P, Kasten, M.M, Ferris, E, Heroux, A, Hill, C.P, Cairns, B.R. | | Deposit date: | 2007-08-21 | | Release date: | 2007-10-30 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation.
Mol.Cell, 27, 2007
|
|
2R3C
 
 | | Structure of the gp41 N-peptide in complex with the HIV entry inhibitor PIE1 | | Descriptor: | CHLORIDE ION, HIV entry inhibitor PIE1, YTTRIUM (III) ION, ... | | Authors: | VanDemark, A.P, Welch, B, Heroux, A, Hill, C.P, Kay, M.S. | | Deposit date: | 2007-08-29 | | Release date: | 2007-10-02 | | Last modified: | 2018-08-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Potent D-peptide inhibitors of HIV-1 entry
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2R5B
 
 | | Structure of the gp41 N-trimer in complex with the HIV entry inhibitor PIE7 | | Descriptor: | HIV entry inhibitor PIE7, SULFATE ION, gp41 N-peptide | | Authors: | VanDemark, A.P, Welch, B, Heroux, A, Hill, C.P, Kay, M.S. | | Deposit date: | 2007-09-03 | | Release date: | 2007-10-02 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent D-peptide inhibitors of HIV-1 entry
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2R5D
 
 | | Structure of the gp41 N-trimer in complex with the HIV entry inhibitor PIE7 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, HIV entry inhibitor PIE7, ... | | Authors: | VanDemark, A.P, Welch, B, Heroux, A, Hill, C.P, Kay, M.S. | | Deposit date: | 2007-09-03 | | Release date: | 2007-10-02 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Potent D-peptide inhibitors of HIV-1 entry
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2RF3
 
 | | Crystal Structure of Tricyclo-DNA: An Unusual Compensatory Change of Two Adjacent Backbone Torsion Angles | | Descriptor: | 5'-d(CGCG(TCY)ATTCGCG)-3', SPERMINE, ZINC ION | | Authors: | Pallan, P.S, Ittig, D, Heroux, A, Wawrzak, Z, Leumann, C.J, Egli, M. | | Deposit date: | 2007-09-27 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of tricyclo-DNA: an unusual compensatory change of two adjacent backbone torsion angles.
Chem.Commun.(Camb.), 2008, 2008
|
|
3RLE
 
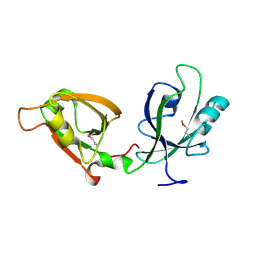 | | Crystal Structure of GRASP55 GRASP domain (residues 7-208) | | Descriptor: | Golgi reassembly-stacking protein 2 | | Authors: | Truschel, S.T, Sengupta, D, Foote, A, Heroux, A, Macbeth, M.R, Linstedt, A.D. | | Deposit date: | 2011-04-19 | | Release date: | 2011-05-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Structure of the Membrane-tethering GRASP Domain Reveals a Unique PDZ Ligand Interaction That Mediates Golgi Biogenesis.
J.Biol.Chem., 286, 2011
|
|
3LX0
 
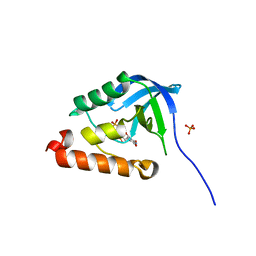 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS D21N at cryogenic temperature | | Descriptor: | PHOSPHATE ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Doctrow, B.M, Schlessman, J.L, Garcia-Moreno, E.B, Heroux, A. | | Deposit date: | 2010-02-24 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cooperative Proton Binding in a Cluster of Carboxylic Residues
To be Published
|
|
4EQN
 
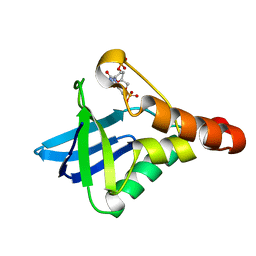 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23E/I72K at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Robinson, A.C, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2012-04-19 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23E/I72K at cryogenic temperature
To be Published
|
|
4EQP
 
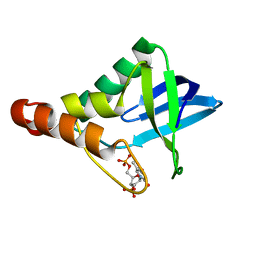 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS I72D at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Robinson, A.C, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2012-04-19 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS I72D at cryogenic temperature
To be Published
|
|
4GG2
 
 | | The crystal structure of glutamate-bound human gamma-glutamyltranspeptidase 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | West, M.B, Chen, Y, Wickham, S, Heroux, A, Cahill, K, Hanigan, M.H, Mooers, B.H.M. | | Deposit date: | 2012-08-04 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Novel Insights into Eukaryotic gamma-Glutamyltranspeptidase 1 from the Crystal Structure of the Glutamate-bound Human Enzyme.
J.Biol.Chem., 288, 2013
|
|
4GDX
 
 | | Crystal Structure of Human Gamma-Glutamyl Transpeptidase--Glutamate complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | West, M.B, Chen, Y, Wickham, S, Heroux, A, Cahill, K, Hanigan, M.H, Mooers, B.H.M. | | Deposit date: | 2012-08-01 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Novel Insights into Eukaryotic gamma-Glutamyltranspeptidase 1 from the Crystal Structure of the Glutamate-bound Human Enzyme.
J.Biol.Chem., 288, 2013
|
|
4HMJ
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L36D at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Robinson, A.C, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2012-10-18 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS L36D at cryogenic temperature
To be Published
|
|
4J2N
 
 | | Crystal Structure of mycobacteriophage Pukovnik Xis | | Descriptor: | Gp37, SULFATE ION | | Authors: | Homa, N.J, Amrich, C.G, Heroux, A, VanDemark, A.P. | | Deposit date: | 2013-02-04 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | The Structure of Xis Reveals the Basis for Filament Formation and Insight into DNA Bending within a Mycobacteriophage Intasome.
J.Mol.Biol., 426, 2014
|
|
4KAP
 
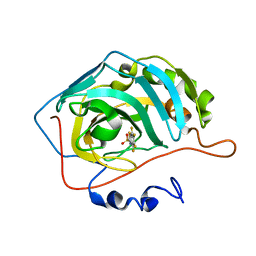 | | The Binding of Benzoarylsulfonamide Ligands to Human Carbonic Anhydrase is Insensitive to Formal Fluorination of the Ligand | | Descriptor: | 4,5,6,7-tetrafluoro-1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Lange, H, Lockett, M.R, Breiten, B, Heroux, A, Sherman, W, Rappoport, D, Yau, P.O, Snyder, P.W, Whitesides, G.M. | | Deposit date: | 2013-04-22 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Binding of Benzoarylsulfonamide Ligands to Human Carbonic Anhydrase is Insensitive to Formal Fluorination of the Ligand.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4L1P
 
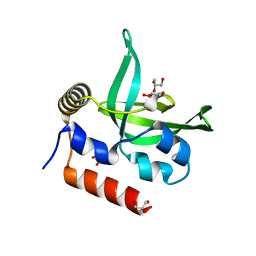 | |
4L2M
 
 | | Crystal structure of the 2/2 hemoglobin from Synechococcus sp. PCC 7002 in the cyanomet state and with covalently attached heme | | Descriptor: | CYANIDE ION, Cyanoglobin, HEME B/C, ... | | Authors: | Wenke, B.B, Schlessman, J.L, Heroux, A, Lecomte, J.T.J. | | Deposit date: | 2013-06-04 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The 2/2 hemoglobin from the cyanobacterium Synechococcus sp. PCC 7002 with covalently attached heme: Comparison of X-ray and NMR structures.
Proteins, 82, 2014
|
|
4KY5
 
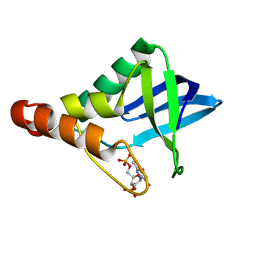 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23D at pH 7 and cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Robinson, A.C, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2013-05-28 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23D at pH 7 and cryogenic temperature
To be Published
|
|
4L1U
 
 | | Crystal Structure of Human Rtf1 Plus3 Domain in Complex with Spt5 CTR Phosphopeptide | | Descriptor: | GLYCEROL, RNA polymerase-associated protein RTF1 homolog, SULFATE ION, ... | | Authors: | Wier, A.D, Heroux, A, VanDemark, A.P. | | Deposit date: | 2013-06-03 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.424 Å) | | Cite: | Structural basis for Spt5-mediated recruitment of the Paf1 complex to chromatin.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4LAA
 
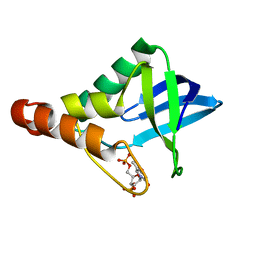 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L36H at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Wheeler, E.L, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B, Robinson, A.C. | | Deposit date: | 2013-06-19 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS L36H at cryogenic temperature
To be Published
|
|
4MAX
 
 | | Crystal structure of Synechococcus sp. PCC 7002 globin at cryogenic temperature with heme modification | | Descriptor: | HEME B/C, SULFATE ION, cyanoglobin | | Authors: | Wenke, B.B, Schlessman, J.L, Heroux, A, Lecomte, J.T.J. | | Deposit date: | 2013-08-18 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.444 Å) | | Cite: | The 2/2 hemoglobin from the cyanobacterium Synechococcus sp. PCC 7002 with covalently attached heme: Comparison of X-ray and NMR structures.
Proteins, 82, 2014
|
|
