2BWN
 
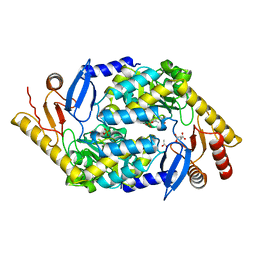 | | 5-Aminolevulinate Synthase from Rhodobacter capsulatus | | Descriptor: | 5-AMINOLEVULINATE SYNTHASE, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Astner, I, Schulze, J.O, van den Heuvel, J.J, Jahn, D, Schubert, W.-D, Heinz, D.W. | | Deposit date: | 2005-07-15 | | Release date: | 2005-09-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of 5-Aminolevulinate Synthase, the First Enzyme of Heme Biosynthesis, and its Link to Xlsa in Humans.
Embo J., 24, 2005
|
|
2C18
 
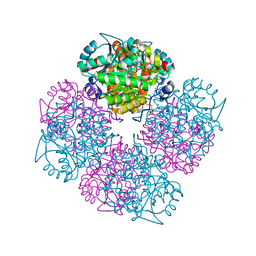 | | 5-(4-Carboxy-2-oxo-butane-1-sulfonyl)-4-oxo-pentanoic acid bound to Porphobilinogen synthase from Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, DELTA-AMINOLEVULINIC ACID DEHYDRATASE, MAGNESIUM ION, ... | | Authors: | Frere, F, Nentwich, M, Gacond, S, Heinz, D.W, Neier, R, Frankenberg-Dinkel, N. | | Deposit date: | 2005-09-11 | | Release date: | 2006-06-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Probing the Active Site of Pseudomonas Aeruginosa Porphobilinogen Synthase Using Newly Developed Inhibitors.
Biochemistry, 45, 2006
|
|
2C4B
 
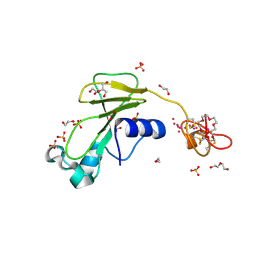 | | Inhibitor cystine knot protein McoEeTI fused to the catalytically inactive barnase mutant H102A | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BARNASE MCOEETI FUSION, ... | | Authors: | Niemann, H.H, Schmoldt, H.U, Wentzel, A, Kolmar, H, Heinz, D.W. | | Deposit date: | 2005-10-18 | | Release date: | 2005-11-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Barnase Fusion as a Tool to Determine the Crystal Structure of the Small Disulfide-Rich Protein Mcoeeti.
J.Mol.Biol., 356, 2006
|
|
2BEO
 
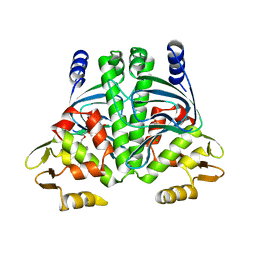 | | PrfA, Transcriptional Regulator In Listeria Monocytogenes | | Descriptor: | ACETAMIDE, CHLORIDE ION, GLUTAMINE, ... | | Authors: | Eiting, M, Hagelueken, G, Schubert, W.-D, Heinz, D.W. | | Deposit date: | 2004-11-26 | | Release date: | 2005-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Mutation G145S in Prfa, a Key Virulence Regulator of Listeria Monocytogenes, Increases DNA-Binding Affinity by Stabilizing the Hth Motif
Mol.Microbiol., 56, 2005
|
|
2C14
 
 | | 5-(4-Carboxy-2-oxo-butylamino)-4-oxo-pentanoic acid acid bound to Porphobilinogen synthase from Pseudomonas aeruginosa | | Descriptor: | DELTA-AMINOLEVULINIC ACID DEHYDRATASE, MAGNESIUM ION | | Authors: | Frere, F, Nentwich, M, Gacond, S, Heinz, D.W, Neier, R, Frankenberg-Dinkel, N. | | Deposit date: | 2005-09-11 | | Release date: | 2006-06-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the Active Site of Pseudomonas Aeruginosa Porphobilinogen Synthase Using Newly Developed Inhibitors.
Biochemistry, 45, 2006
|
|
2C19
 
 | | 5-(4-Carboxy-2-oxo-butylsulfanyl)-4-oxo-pentanoic acid acid bound to Porphobilinogen synthase from Pseudomonas aeruginosa | | Descriptor: | DELTA-AMINOLEVULINIC ACID DEHYDRATASE, MAGNESIUM ION, SODIUM ION | | Authors: | Frere, F, Nentwich, M, Gacond, S, Heinz, D.W, Neier, R, Frankenberg-Dinkel, N. | | Deposit date: | 2005-09-11 | | Release date: | 2006-06-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Probing the Active Site of Pseudomonas Aeruginosa Porphobilinogen Synthase Using Newly Developed Inhibitors.
Biochemistry, 45, 2006
|
|
2CG3
 
 | | Crystal structure of SdsA1, an alkylsulfatase from Pseudomonas aeruginosa. | | Descriptor: | SDSA1, ZINC ION | | Authors: | Hagelueken, G, Adams, T.M, Wiehlmann, L, Widow, U, Kolmar, H, Tuemmler, B, Heinz, D.W, Schubert, W.-D. | | Deposit date: | 2006-02-27 | | Release date: | 2006-04-26 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of Sdsa1, an Alkylsulfatase from Pseudomonas Aeruginosa, Defines a Third Third Class of Sulfatases
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2CIA
 
 | | human nck2 sh2-domain in complex with a decaphosphopeptide from translocated intimin receptor (tir) of epec | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CYTOPLASMIC PROTEIN NCK2, ISOPROPYL ALCOHOL, ... | | Authors: | Frese, S, Schubert, W.-D, Findeis, A.C, Marquardt, T, Roske, Y.S, Stradal, T.E.B, Heinz, D.W. | | Deposit date: | 2006-03-17 | | Release date: | 2006-04-24 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Phosphotyrosine Peptide Binding Specificity of Nck1 and Nck2 Src Homology 2 Domains.
J.Biol.Chem., 281, 2006
|
|
2CI8
 
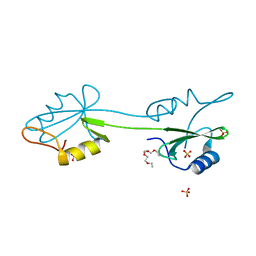 | | sh2 domain of human nck1 adaptor protein - uncomplexed | | Descriptor: | CYTOPLASMIC PROTEIN NCK1, PENTAETHYLENE GLYCOL, SULFATE ION | | Authors: | Frese, S, Schubert, W.-D, Findeis, A.C, Marquardt, T, Roske, Y.S, Stradal, T.E.B, Heinz, D.W. | | Deposit date: | 2006-03-17 | | Release date: | 2006-04-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Phosphotyrosine Peptide Binding Specificity of Nck1 and Nck2 Src Homology 2 Domains.
J.Biol.Chem., 281, 2006
|
|
2CI9
 
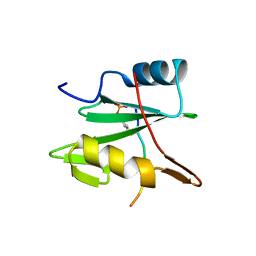 | | Nck1 SH2-domain in complex with a dodecaphosphopeptide from EPEC protein Tir | | Descriptor: | CYTOPLASMIC PROTEIN NCK1, TRANSLOCATED INTIMIN RECEPTOR | | Authors: | Frese, S, Schubert, W.-D, Findeis, A.C, Marquardt, T, Roske, Y.S, Stradal, T.E.B, Heinz, D.W. | | Deposit date: | 2006-03-17 | | Release date: | 2006-04-24 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Phosphotyrosine Peptide Binding Specificity of Nck1 and Nck2 Src Homology 2 Domains.
J.Biol.Chem., 281, 2006
|
|
2CG2
 
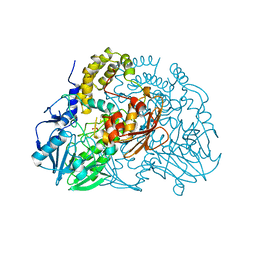 | | Crystal structure of SdsA1, an alkylsulfatase from Pseudomonas aeruginosa, in complex with sulfate | | Descriptor: | SDSA1, SULFATE ION, ZINC ION | | Authors: | Hagelueken, G, Adams, T.M, Wiehlmann, L, Widow, U, Kolmar, H, Tuemmler, B, Heinz, D.W, Schubert, W.-D. | | Deposit date: | 2006-02-27 | | Release date: | 2006-04-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of Sdsa1, an Alkylsulfatase from Pseudomonas Aeruginosa, Defines a Third Class of Sulfatases.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2CFO
 
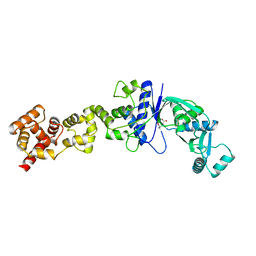 | | Non-Discriminating Glutamyl-tRNA Synthetase from Thermosynechococcus elongatus in Complex with Glu | | Descriptor: | GLUTAMIC ACID, GLUTAMYL-TRNA SYNTHETASE | | Authors: | Schulze, J.O, Nickel, D, Schubert, W.-D, Jahn, D, Heinz, D.W. | | Deposit date: | 2006-02-22 | | Release date: | 2006-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of a Non-Discriminating Glutamyl- tRNA Synthetase.
J.Mol.Biol., 361, 2006
|
|
2CFZ
 
 | | Crystal structure of SdsA1, an alkylsulfatase from Pseudomonas aeruginosa, in complex with 1-dodecanol | | Descriptor: | 1-DODECANOL, DI(HYDROXYETHYL)ETHER, SDS HYDROLASE SDSA1, ... | | Authors: | Hagelueken, G, Adams, T.M, Wiehlmann, L, Widow, U, Kolmar, H, Tuemmler, B, Heinz, D.W, Schubert, W.-D. | | Deposit date: | 2006-02-26 | | Release date: | 2006-04-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Crystal Structure of Sdsa1, an Alkylsulfatase from Pseudomonas Aeruginosa, Defines a Third Class of Sulfatases.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2CFU
 
 | | Crystal structure of SdsA1, an alkylsulfatase from Pseudomonas aeruginosa, in complex with 1-decane-sulfonic-acid. | | Descriptor: | 1-DECANE-SULFONIC-ACID, DI(HYDROXYETHYL)ETHER, ISOPROPYL ALCOHOL, ... | | Authors: | Hagelueken, G, Adams, T.M, Wiehlmann, L, Widow, U, Kolmar, H, Tuemmler, B, Heinz, D.W, Schubert, W.-D. | | Deposit date: | 2006-02-23 | | Release date: | 2006-04-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of Sdsa1, an Alkylsulfatase from Pseudomonas Aeruginosa, Defines a Third Class of Sulfatases.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2CFB
 
 | | Glutamate-1-semialdehyde 2,1-Aminomutase from Thermosynechococcus elongatus | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, GLUTAMATE-1-SEMIALDEHYDE 2,1-AMINOMUTASE | | Authors: | Schulze, J.O, Schubert, W.-D, Moser, J, Jahn, D, Heinz, D.W. | | Deposit date: | 2006-02-17 | | Release date: | 2006-03-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Evolutionary Relationship between Initial Enzymes of Tetrapyrrole Biosynthesis
J.Mol.Biol., 358, 2006
|
|
1GZG
 
 | | Complex of a Mg2-dependent porphobilinogen synthase from Pseudomonas aeruginosa (mutant D139N) with 5-fluorolevulinic acid | | Descriptor: | 5-FLUOROLEVULINIC ACID, DELTA-AMINOLEVULINIC ACID DEHYDRATASE, MAGNESIUM ION, ... | | Authors: | Frere, F, Schubert, W.-D, Stauffer, F, Frankenberg, N, Neier, R, Jahn, D, Heinz, D.W. | | Deposit date: | 2002-05-21 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure of porphobilinogen synthase from Pseudomonas aeruginosa in complex with 5-fluorolevulinic acid suggests a double Schiff base mechanism.
J. Mol. Biol., 320, 2002
|
|
1I7A
 
 | | EVH1 DOMAIN FROM MURINE HOMER 2B/VESL 2 | | Descriptor: | CITRATE ANION, HOMER 2B, PHE-ALA-PHE, ... | | Authors: | Barzik, M, Carl, U.D, Schubert, W.-D, Wehland, J, Heinz, D.W. | | Deposit date: | 2001-03-08 | | Release date: | 2001-08-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The N-terminal domain of Homer/Vesl is a new class II EVH1 domain.
J.Mol.Biol., 309, 2001
|
|
1L64
 
 | |
1L67
 
 | |
1L68
 
 | |
1L66
 
 | |
1L65
 
 | |
6LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2021-06-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
4LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
5LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2021-06-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
