5KXC
 
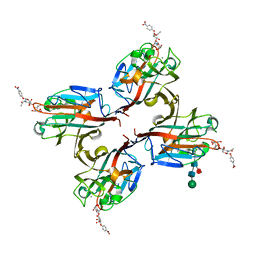 | | Wisteria floribunda lectin in complex with GalNAc(beta1-4)GlcNAc (LacdiNAc) at pH 8.5. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Evans, S.V, Haji-Ghassemi, O. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis for Recognition of the Cancer Glycobiomarker, LacdiNAc (GalNAc[ beta 14]GlcNAc), by Wisteria floribunda Agglutinin.
J.Biol.Chem., 291, 2016
|
|
5KXB
 
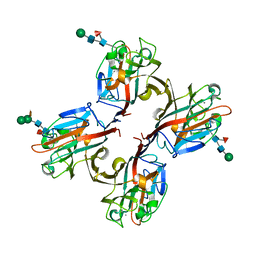 | | Wisteria floribunda lectin in complex with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Evans, S.V, Haji-Ghassemi, O. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Molecular Basis for Recognition of the Cancer Glycobiomarker, LacdiNAc (GalNAc[ beta 14]GlcNAc), by Wisteria floribunda Agglutinin.
J.Biol.Chem., 291, 2016
|
|
5KXE
 
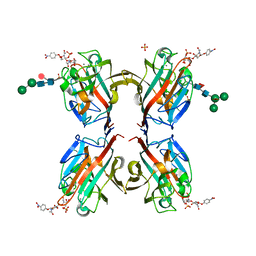 | | Wisteria floribunda lectin in complex with GalNAc(beta1-4)GlcNAc (LacdiNAc) at pH 4.2 | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Evans, S.V, Haji-Ghassemi, O. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Molecular Basis for Recognition of the Cancer Glycobiomarker, LacdiNAc (GalNAc[ beta 14]GlcNAc), by Wisteria floribunda Agglutinin.
J.Biol.Chem., 291, 2016
|
|
6C5I
 
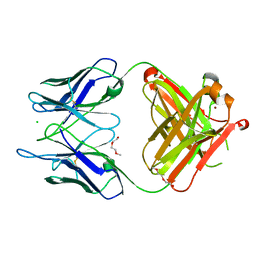 | | Unliganded S25-5 Fab | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Evans, S.V, Haji-Ghassemi, O. | | Deposit date: | 2018-01-16 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Subtle Changes in the Combining Site of the Chlamydiaceae-Specific mAb S25-23 Increase the Antibody-Carbohydrate Binding Affinity by an Order of Magnitude.
Biochemistry, 58, 2019
|
|
6C5H
 
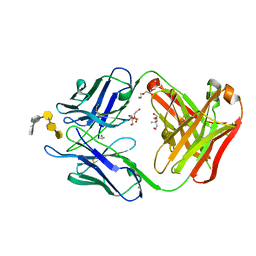 | | S25-5 Fab in complex with Chlamydiaceae-specific LPS antigen | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-8)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)-2-amino-2-deoxy-4-O-phosphono-beta-D-glucopyranose-(1-6)-2-amino-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Evans, S.V, Haji-Ghassemi, O. | | Deposit date: | 2018-01-16 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Subtle Changes in the Combining Site of the Chlamydiaceae-Specific mAb S25-23 Increase the Antibody-Carbohydrate Binding Affinity by an Order of Magnitude.
Biochemistry, 58, 2019
|
|
6W1N
 
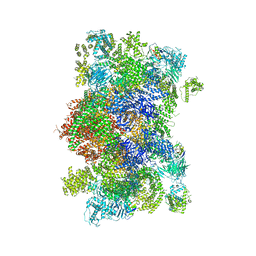 | |
7CF9
 
 | | Structure of RyR1 (Ca2+/CHL) | | Descriptor: | 5-bromanyl-N-[4-chloranyl-2-methyl-6-(methylcarbamoyl)phenyl]-2-(3-chloranylpyridin-2-yl)pyrazole-3-carboxamide, CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ... | | Authors: | Ma, R, Haji-Ghassemi, O, Ma, D, Lin, L, Samurkas, A, Van Petegem, F, Yuchi, Z. | | Deposit date: | 2020-06-24 | | Release date: | 2020-09-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural basis for diamide modulation of ryanodine receptor.
Nat.Chem.Biol., 16, 2020
|
|
6M2W
 
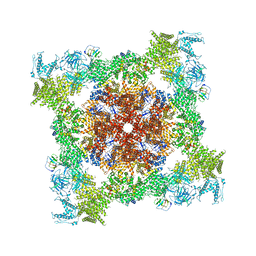 | | Structure of RyR1 (Ca2+/Caffeine/ATP/CaM1234/CHL) | | Descriptor: | 5-bromanyl-N-[4-chloranyl-2-methyl-6-(methylcarbamoyl)phenyl]-2-(3-chloranylpyridin-2-yl)pyrazole-3-carboxamide, ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, ... | | Authors: | Ma, R, Haji-Ghassemi, O, Ma, D, Lin, L, Samurkas, A, Van Petegem, F, Yuchi, Z. | | Deposit date: | 2020-03-01 | | Release date: | 2020-09-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for diamide modulation of ryanodine receptor.
Nat.Chem.Biol., 16, 2020
|
|
6X35
 
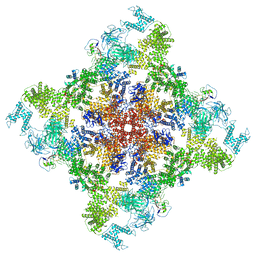 | | Pig R615C RyR1 in complex with CaM, EGTA (class 1, open) | | Descriptor: | Calmodulin-1, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine Receptor, ... | | Authors: | Woll, K.W, Haji-Ghassemi, O, Van Petegem, F. | | Deposit date: | 2020-05-21 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Pathological conformations of disease mutant Ryanodine Receptors revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
6X33
 
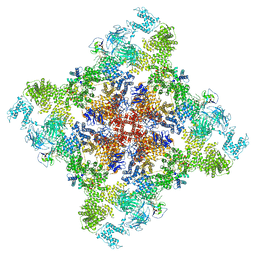 | | Wt pig RyR1 in complex with apoCaM, EGTA condition (class 3, open) | | Descriptor: | Calmodulin-1, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine Receptor, ... | | Authors: | Woll, K.W, Haji-Ghassemi, O, Van Petegem, F. | | Deposit date: | 2020-05-21 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Pathological conformations of disease mutant Ryanodine Receptors revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
6X32
 
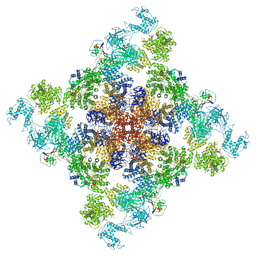 | | Wt pig RyR1 in complex with apoCaM, EGTA condition (class 1 and 2, closed) | | Descriptor: | Calmodulin-1, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine Receptor, ... | | Authors: | Woll, K.W, Haji-Ghassemi, O, Van Petegem, F. | | Deposit date: | 2020-05-21 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Pathological conformations of disease mutant Ryanodine Receptors revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
6X34
 
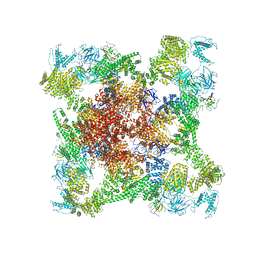 | | Pig R615C RyR1 EGTA (all classes, open) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine Receptor, ZINC ION | | Authors: | Woll, K.W, Haji-Ghassemi, O, Van Petegem, F. | | Deposit date: | 2020-05-21 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Pathological conformations of disease mutant Ryanodine Receptors revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
6X36
 
 | | Pig R615C RyR1 in complex with CaM, EGTA (class 3, closed) | | Descriptor: | Calmodulin-1, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine Receptor | | Authors: | Woll, K.W, Haji-Ghassemi, O, Van Petegem, F. | | Deposit date: | 2020-05-21 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Pathological conformations of disease mutant Ryanodine Receptors revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
6CWR
 
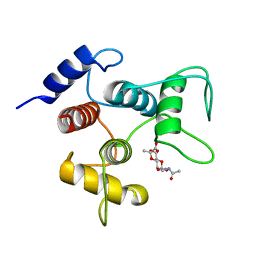 | | Crystal structure of SpaA-SLH/G46A/G109A in complex with 4,6-Pyr-beta-D-ManNAcOMe | | Descriptor: | Surface (S-) layer glycoprotein, methyl 2-(acetylamino)-4,6-O-[(1S)-1-carboxyethylidene]-2-deoxy-beta-D-mannopyranoside | | Authors: | Blackler, R.J, Gagnon, S.M.L, Haji-Ghassemi, O, Evans, S.V. | | Deposit date: | 2018-03-30 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structural basis of cell wall anchoring by SLH domains in Paenibacillus alvei.
Nat Commun, 9, 2018
|
|
8SEU
 
 | | Cryo-EM Structure of RyR1 (Local Refinement of TMD) | | Descriptor: | Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEP
 
 | | Cryo-EM Structure of RyR1 + ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SER
 
 | | Cryo-EM Structure of RyR1 + Adenosine | | Descriptor: | ADENOSINE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEV
 
 | | Cryo-EM Structure of RyR1 + ATP-gamma-S (Local Refinement of TMD) | | Descriptor: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEY
 
 | | Cryo-EM Structure of RyR1 + Adenosine (Local Refinement of TMD) | | Descriptor: | ADENOSINE, Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEQ
 
 | | Cryo-EM Structure of RyR1 + AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEW
 
 | | Cryo-EM Structure of RyR1 + ADP (Local Refinement of TMD) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SF0
 
 | | Cryo-EM Structure of RyR1 + cAMP (Local Refinement of TMD) | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SES
 
 | | Cryo-EM Structure of RyR1 + Adenine | | Descriptor: | ADENINE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEO
 
 | | Cryo-EM Structure of RyR1 + ATP-gamma-S | | Descriptor: | Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SET
 
 | | Cryo-EM Structure of RyR1 + cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
