7V6Q
 
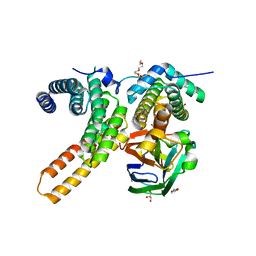 | | Crystal structure of sNASP-ASF1A-H3.1-H4 complex | | 分子名称: | GLYCEROL, Histone H3.1, Histone H4, ... | | 著者 | Liu, C.P, Xu, R.M. | | 登録日 | 2021-08-20 | | 公開日 | 2021-12-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Distinct histone H3-H4 binding modes of sNASP reveal the basis for cooperation and competition of histone chaperones.
Genes Dev., 35, 2021
|
|
7DLA
 
 | |
7DL9
 
 | |
7CYP
 
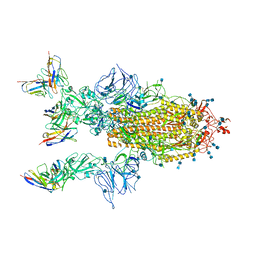 | | Complex of SARS-CoV-2 spike trimer with its neutralizing antibody HB27 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of HB27, ... | | 著者 | Wang, X, Zhu, L. | | 登録日 | 2020-09-04 | | 公開日 | 2021-06-09 | | 最終更新日 | 2022-02-23 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Double lock of a potent human therapeutic monoclonal antibody against SARS-CoV-2.
Natl Sci Rev, 8, 2021
|
|
7VQQ
 
 | |
7VPN
 
 | |
7W5P
 
 | | Crystal Structure of the dioxygenase CcTet from Coprinopsis cinereain bound to 12bp N6-methyldeoxyadenine (6mA) containing duplex DNA | | 分子名称: | CcTet, DNA, DNA (12-MER), ... | | 著者 | Mu, Y.J, Zhang, L, Zhang, L. | | 登録日 | 2021-11-30 | | 公開日 | 2022-03-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | A fungal dioxygenase CcTet serves as a eukaryotic 6mA demethylase on duplex DNA.
Nat.Chem.Biol., 18, 2022
|
|
7ELG
 
 | | LC3B modificated with a covalent probe | | 分子名称: | 2-methylidene-5-thiophen-2-yl-cyclohexane-1,3-dione, Microtubule-associated proteins 1A/1B light chain 3B, SULFATE ION | | 著者 | Fan, S, Wan, W. | | 登録日 | 2021-04-10 | | 公開日 | 2021-10-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.599 Å) | | 主引用文献 | Inhibition of Autophagy by a Small Molecule through Covalent Modification of the LC3 Protein.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
2KQT
 
 | | Solid-state NMR structure of the M2 transmembrane peptide of the influenza A virus in DMPC lipid bilayers bound to deuterated amantadine | | 分子名称: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, M2 protein | | 著者 | Cady, S.D, Schmidt-Rohr, K, Wang, J, Soto, C.S, DeGrado, W.F, Hong, M. | | 登録日 | 2009-11-18 | | 公開日 | 2010-02-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers
Nature, 463, 2010
|
|
5TEC
 
 | | Crystal structure of the TIR domain from the Arabidopsis thaliana NLR protein SNC1 | | 分子名称: | Protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1 | | 著者 | Zhang, X, Bentham, A, Ve, T, Williams, S.J, Kobe, B. | | 登録日 | 2016-09-20 | | 公開日 | 2017-02-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Multiple functional self-association interfaces in plant TIR domains.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5TEB
 
 | | Crystal Structure of the TIR domain from the Arabidopsis Thaliana disease resistance protein RPP1 | | 分子名称: | Recognition of Peronospora parasitica 1 | | 著者 | Bentham, A.R, Zhang, X, Croll, T, Williams, S, Kobe, B. | | 登録日 | 2016-09-20 | | 公開日 | 2017-02-01 | | 最終更新日 | 2020-01-01 | | 実験手法 | X-RAY DIFFRACTION (2.796 Å) | | 主引用文献 | Multiple functional self-association interfaces in plant TIR domains.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VXO
 
 | |
5VXS
 
 | |
5VXC
 
 | |
7VS2
 
 | |
7XH6
 
 | | Crystal structure of CBP bromodomain liganded with CCS1477 | | 分子名称: | (6S)-1-[3,4-bis(fluoranyl)phenyl]-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-(4-methoxycyclohexyl)benzimidazol-2-yl]piperidin-2-one, CREB-binding protein, DIMETHYL SULFOXIDE, ... | | 著者 | Xu, H, Xiang, Q, Wang, C, Zhang, C, Luo, G, Wu, X, Zhang, Y, Xu, Y. | | 登録日 | 2022-04-07 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural insights revealed by the cocrystal structure of CCS1477 in complex with CBP bromodomain
Biochem.Biophys.Res.Commun., 623, 2022
|
|
7XI0
 
 | | Crystal structure of CBP bromodomain liganded with CCS150 | | 分子名称: | (6S)-1-(3-chloranyl-4-methoxy-phenyl)-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(3R)-1-methylsulfonylpyrrolidin-3-yl]benzimidazol-2-yl]piperidin-2-one, CREB-binding protein, GLYCEROL | | 著者 | Xu, H, Xiang, Q, Wang, C, Zhang, C, Luo, G, Wu, X, Zhang, Y, Xu, Y. | | 登録日 | 2022-04-11 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Structural insights revealed by the cocrystal structure of CCS1477 in complex with CBP bromodomain
Biochem.Biophys.Res.Commun., 623, 2022
|
|
7XHE
 
 | | Crystal structure of CBP bromodomain liganded with CCS151 | | 分子名称: | (6S)-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(3R)-1-methylsulfonylpyrrolidin-3-yl]benzimidazol-2-yl]-1-(3-fluoranyl-4-methoxy-phenyl)piperidin-2-one, 1,2-ETHANEDIOL, CREB-binding protein | | 著者 | Xu, H, Xiang, Q, Wang, C, Zhang, C, Luo, G, Wu, X, Zhang, Y, Xu, Y. | | 登録日 | 2022-04-08 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Structural insights revealed by the cocrystal structure of CCS1477 in complex with CBP bromodomain
Biochem.Biophys.Res.Commun., 623, 2022
|
|
5YEW
 
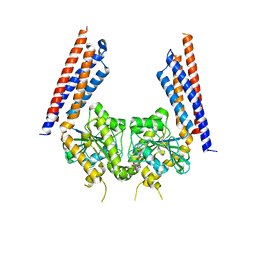 | | Structural basis for GTP hydrolysis and conformational change of mitofusin 1 in mediating mitochondrial fusion | | 分子名称: | BERYLLIUM TRIFLUORIDE ION, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Yan, L, Qi, Y, Huang, X, Yu, C. | | 登録日 | 2017-09-20 | | 公開日 | 2018-01-31 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural basis for GTP hydrolysis and conformational change of MFN1 in mediating membrane fusion
Nat. Struct. Mol. Biol., 25, 2018
|
|
5YWO
 
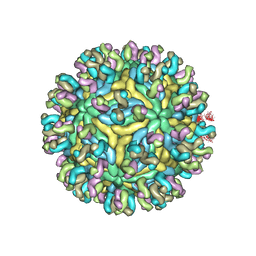 | | Structure of JEV-2F2 Fab complex | | 分子名称: | 2F2 heavy chain, 2F2 light chain, JEV E protein, ... | | 著者 | Qiu, X, Lei, Y.F, Yang, P, Gao, Q, Wang, N, Cao, L, Yuan, S, Wang, X, Xu, Z.K, Rao, Z. | | 登録日 | 2017-11-29 | | 公開日 | 2018-03-21 | | 最終更新日 | 2018-09-12 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Structural basis for neutralization of Japanese encephalitis virus by two potent therapeutic antibodies
Nat Microbiol, 3, 2018
|
|
5YWP
 
 | | JEV-2H4 Fab complex | | 分子名称: | 2H4 heavy chain, 2H4 light chain, JEV E protein, ... | | 著者 | Qiu, X.D, Lei, Y.F, Yang, P, Gao, Q, WANG, N, Cao, L, Yuan, S, Wang, X.X, Xu, Z.K, Rao, Z.H. | | 登録日 | 2017-11-29 | | 公開日 | 2018-05-02 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Structural basis for neutralization of Japanese encephalitis virus by two potent therapeutic antibodies
Nat Microbiol, 3, 2018
|
|
5YWF
 
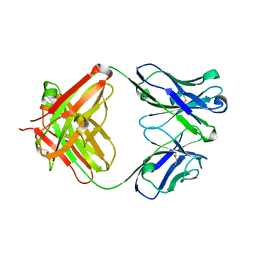 | | Crystal structure of 2H4 Fab | | 分子名称: | 2H4 heavy chain, 2H4 light chain | | 著者 | Qiu, X, Lei, Y, Yang, P, Gao, Q, Wang, N, Cao, L, Wang, X, Xu, Z.K, Rao, Z. | | 登録日 | 2017-11-29 | | 公開日 | 2018-03-21 | | 実験手法 | X-RAY DIFFRACTION (2.206 Å) | | 主引用文献 | Structural basis for neutralization of Japanese encephalitis virus by two potent therapeutic antibodies
Nat Microbiol, 3, 2018
|
|
5GNT
 
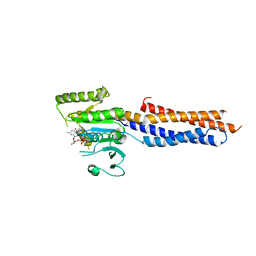 | | BDLP-like folding of Mitofusin 1 | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, Mitofusin-1 | | 著者 | Yan, L, Yu, C, Ming, Z, Rao, Z, Lou, Z, Hu, J. | | 登録日 | 2016-07-24 | | 公開日 | 2016-11-23 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.665 Å) | | 主引用文献 | the structure of mini-MFN1 in complex with GDP
To Be Published
|
|
7JKB
 
 | | 2xVH Fab | | 分子名称: | Anti-Her2, Anti-lysozyme | | 著者 | Lord, D.M, Zhou, Y.F. | | 登録日 | 2020-07-28 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Bringing the Heavy Chain to Light: Creating a Symmetric, Bivalent IgG-Like Bispecific.
Antibodies, 9, 2020
|
|
6DG3
 
 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase, in complex with caesium | | 分子名称: | CESIUM ION, PHOSPHATE ION, Pyridinium-3,5-biscarboxylic acid mononucleotide sulfurtransferase | | 著者 | Fellner, M, Hausinger, R.P, Hu, J. | | 登録日 | 2018-05-16 | | 公開日 | 2019-05-22 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.944 Å) | | 主引用文献 | Unique cesium-binding sites in proteins, a case study with the sacrificial sulfur transferase LarE
J Life Sci, 2021
|
|
