3J2F
 
 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (17.6 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process.
Nucleic Acids Res., 41, 2013
|
|
3J2H
 
 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (18.799999 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process.
Nucleic Acids Res., 41, 2013
|
|
3J2B
 
 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (13.6 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process
Nucleic Acids Res., 41, 2013
|
|
3J2A
 
 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (13.1 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process
Nucleic Acids Res., 41, 2013
|
|
3KBX
 
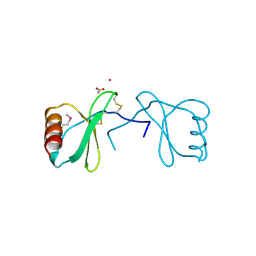 | |
5YBJ
 
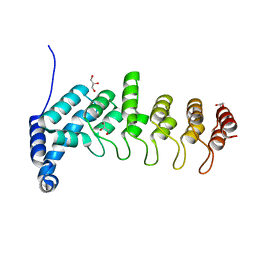 | | Structure of apo KANK1 ankyrin domain | | Descriptor: | GLYCEROL, KN motif and ankyrin repeat domain-containing protein 1 | | Authors: | Guo, Q, Liao, S, Min, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-05 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | Structural basis for the recognition of kinesin family member 21A (KIF21A) by the ankyrin domains of KANK1 and KANK2 proteins.
J. Biol. Chem., 293, 2018
|
|
5YBV
 
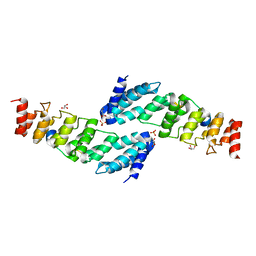 | | The structure of the KANK2 ankyrin domain with the KIF21A peptide | | Descriptor: | GLYCEROL, KN motif and ankyrin repeat domain-containing protein 2, Kinesin-like protein KIF21A, ... | | Authors: | Guo, Q, Liao, S, Min, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-05 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural basis for the recognition of kinesin family member 21A (KIF21A) by the ankyrin domains of KANK1 and KANK2 proteins.
J. Biol. Chem., 293, 2018
|
|
5YBU
 
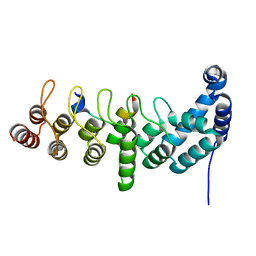 | | Structure of the KANK1 ankyrin domain in complex with KIF21A peptide | | Descriptor: | KN motif and ankyrin repeat domain-containing protein 1, Kinesin-like protein KIF21A | | Authors: | Guo, Q, Liao, S, Min, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-05 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for the recognition of kinesin family member 21A (KIF21A) by the ankyrin domains of KANK1 and KANK2 proteins.
J. Biol. Chem., 293, 2018
|
|
7XDT
 
 | |
7XGR
 
 | |
6ICT
 
 | | Structure of SETD3 bound to SAH and methylated actin | | Descriptor: | Actin, cytoplasmic 1, Histone-lysine N-methyltransferase setd3, ... | | Authors: | Guo, Q, Liao, S, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-07 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Structural insights into SETD3-mediated histidine methylation on beta-actin.
Elife, 8, 2019
|
|
6ICV
 
 | | Structure of SETD3 bound to SAH and unmodified actin | | Descriptor: | Actin, cytoplasmic 1, Histone-lysine N-methyltransferase setd3, ... | | Authors: | Guo, Q, Liao, S, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-07 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into SETD3-mediated histidine methylation on beta-actin.
Elife, 8, 2019
|
|
6K0X
 
 | |
2X6L
 
 | |
2X69
 
 | |
2X6G
 
 | |
2YPU
 
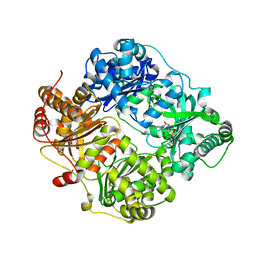 | | human insulin degrading enzyme E111Q in complex with inhibitor compound 41367 | | Descriptor: | 2-[[2-[[(2S)-3-(3H-IMIDAZOL-4-YL)-1-METHOXY-1-OXO-PROPAN-2-YL]AMINO]-2-OXO-ETHYL]-(PHENYLMETHYL)AMINO]ETHANOIC ACID, INSULIN-DEGRADING ENZYME, ZINC ION | | Authors: | Guo, Q, Deprez-Poulain, R, Deprez, B, Tang, W.-J. | | Deposit date: | 2012-11-01 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Imidazole-Derived 2-[N-Carbamoylmethyl-Alkylamino]Acetic Acids,Substrate-Dependent Modulators of Insulin-Degrading Enzyme in Amyloid-Beta Hydrolysis
Eur J Med Chem, 79C, 2014
|
|
4DWK
 
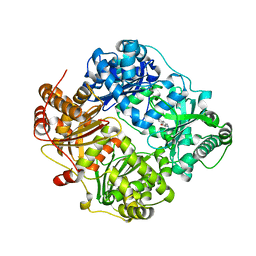 | | Structure of cystein free insulin degrading enzyme with compound bdm41671 ((s)-2-{2-[carboxymethyl-(3-phenyl-propyl)-amino]-acetylamino}-3-(1h-imidazol-4-yl)-propionic acid methyl ester) | | Descriptor: | Insulin-degrading enzyme, ZINC ION, methyl N-(carboxymethyl)-N-(3-phenylpropyl)glycyl-L-histidinate | | Authors: | Guo, Q, Deprez-Poulain, R, Deprez, B, Tang, W.J. | | Deposit date: | 2012-02-24 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Imidazole-derived 2-[N-carbamoylmethyl-alkylamino]acetic acids, substrate-dependent modulators of insulin-degrading enzyme in amyloid-beta hydrolysis.
Eur.J.Med.Chem., 79, 2014
|
|
4DTT
 
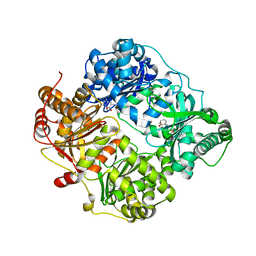 | | Crystal structure of human insulin degrading enzyme (ide) in complex with compund 41367 | | Descriptor: | 2-[[2-[[(2S)-3-(3H-IMIDAZOL-4-YL)-1-METHOXY-1-OXO-PROPAN-2-YL]AMINO]-2-OXO-ETHYL]-(PHENYLMETHYL)AMINO]ETHANOIC ACID, Insulin-degrading enzyme, ZINC ION | | Authors: | Guo, Q, Deprez-Poulain, R, Deprez, B, Tang, W.J. | | Deposit date: | 2012-02-21 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Imidazole-derived 2-[N-carbamoylmethyl-alkylamino]acetic acids, substrate-dependent modulators of insulin-degrading enzyme in amyloid-beta hydrolysis.
Eur.J.Med.Chem., 79, 2014
|
|
6D4B
 
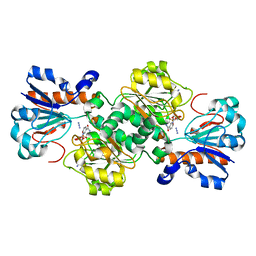 | | Crystal structure of Candida boidinii formate dehydrogenase V123A mutant complexed with NAD+ and azide | | Descriptor: | AZIDE ION, CHLORIDE ION, Formate dehydrogenase, ... | | Authors: | Guo, Q, Ye, H, Gakhar, L, Cheatum, C.M, Kohen, A. | | Deposit date: | 2018-04-17 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Oscillatory Active-site Motions Correlate with Kinetic Isotope Effects in Formate Dehydrogenase
Acs Catalysis, 2019
|
|
6D4C
 
 | | Crystal structure of Candida boidinii formate dehydrogenase V123G mutant complexed with NAD+ and azide | | Descriptor: | AZIDE ION, CHLORIDE ION, Formate dehydrogenase, ... | | Authors: | Guo, Q, Ye, H, Gakhar, L, Cheatum, C.M, Kohen, A. | | Deposit date: | 2018-04-17 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Oscillatory Active-site Motions Correlate with Kinetic Isotope Effects in Formate Dehydrogenase
Acs Catalysis, 2019
|
|
3QZ2
 
 | | The structure of cysteine-free human insulin degrading enzyme | | Descriptor: | Insulin-degrading enzyme, ZINC ION | | Authors: | Guo, Q, Tang, W.J. | | Deposit date: | 2011-03-04 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Imidazole-derived 2-[N-carbamoylmethyl-alkylamino]acetic acids, substrate-dependent modulators of insulin-degrading enzyme in amyloid-beta hydrolysis.
Eur.J.Med.Chem., 79, 2014
|
|
8JMT
 
 | | Structure of the adhesion GPCR ADGRL3 in the apo state | | Descriptor: | Adhesion G protein-coupled receptor L3,Soluble cytochrome b562 | | Authors: | Tao, Y, Guo, Q, He, B, Zhong, Y. | | Deposit date: | 2023-06-05 | | Release date: | 2023-09-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A method for structure determination of GPCRs in various states.
Nat.Chem.Biol., 20, 2024
|
|
4KR1
 
 | |
4GS8
 
 | | Structure analysis of cysteine free insulin degrading enzyme (ide) with compound bdm43079 [{[(s)-2-(1h-imidazol-4-yl)-1-methylcarbamoyl-ethylcarbamoyl]-methyl}-(3-phenyl-propyl)-amino]-acetic acid | | Descriptor: | Insulin-degrading enzyme, N-(carboxymethyl)-N-(3-phenylpropyl)glycyl-N-methyl-L-histidinamide, ZINC ION | | Authors: | Guo, Q, Deprez-Poulain, R, Deprez, B, Tang, W.J. | | Deposit date: | 2012-08-27 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Imidazole-derived 2-[N-carbamoylmethyl-alkylamino]acetic acids, substrate-dependent modulators of insulin-degrading enzyme in amyloid-beta hydrolysis.
Eur.J.Med.Chem., 79, 2014
|
|
