7Q7F
 
 | | Room temperature structure of the Rhodobacter Sphaeroides Photosynthetic Reaction Center F(M197)H mutant at atmospheric pressure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Lieske, J, Guenther, S, Saouane, S, Selikhanov, G.K, Gabdulkhakov, A.G, Meents, A. | | Deposit date: | 2021-11-09 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Fixed-target high-pressure macromolecular crystallography
To Be Published
|
|
7PUX
 
 | | Structure of p97 N-D1(L198W) in complex with Fragment TROLL2 | | Descriptor: | (1S)-2-amino-1-(4-bromophenyl)ethan-1-ol, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bothe, S, Schindelin, H. | | Deposit date: | 2021-10-01 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Fragment screening using biolayer interferometry reveals ligands targeting the SHP-motif binding site of the AAA+ ATPase p97
Commun Chem, 5, 2022
|
|
7QIV
 
 | | Structure of human C3b in complex with the EWE nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3 beta chain, Complement C3b alpha' chain, ... | | Authors: | Pedersen, H, Andersen, G.R. | | Deposit date: | 2021-12-16 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Guided Engineering of a Complement Component C3-Binding Nanobody Improves Specificity and Adds Cofactor Activity.
Front Immunol, 13, 2022
|
|
7QYH
 
 | |
6ZEX
 
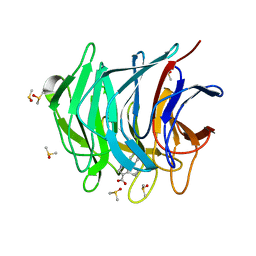 | | Keap1 kelch domain bound to a small molecule fragment | | Descriptor: | 5-cyclopropyl-1-phenyl-pyrazole-4-carboxylic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1 | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
6ZEZ
 
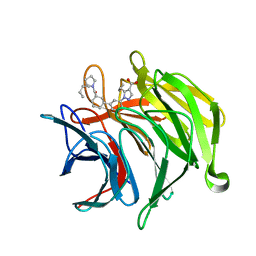 | |
6ZF5
 
 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | Descriptor: | 1-[3-[(4-butylphenyl)sulfonyl-(2-hydroxy-2-oxoethyl)amino]phenyl]-5-cyclopropyl-pyrazole-4-carboxylic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
6Z2H
 
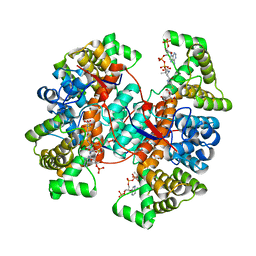 | |
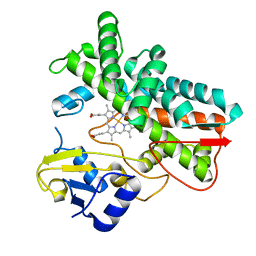
7ABB
 
 | |
7A1A
 
 | |
7S8W
 
 | | Amycolatopsis sp. T-1-60 N-succinylamino acid racemase/o-succinylbenzoate synthase R266Q mutant in complex with N-succinylphenylglycine | | Descriptor: | MAGNESIUM ION, N-succinyl-L-phenylglycine, N-succinylamino acid racemase/O-succinylbenzoate synthase, ... | | Authors: | Truong, D.P, Rousseau, S, Sacchettini, J.C, Glasner, M.E. | | Deposit date: | 2021-09-20 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Second-Shell Amino Acid R266 Helps Determine N -Succinylamino Acid Racemase Reaction Specificity in Promiscuous N -Succinylamino Acid Racemase/ o -Succinylbenzoate Synthase Enzymes.
Biochemistry, 60, 2021
|
|
7SDP
 
 | |
7SOZ
 
 | | Replication Initiator Protein REPE54 and cognate DNA sequence with terminal three prime phosphates chemically crosslinked (5 mg/mL EDC, 12 hours). | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*A)-3'), DNA (5'-D(*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*A)-3'), MAGNESIUM ION, ... | | Authors: | Ward, A.R, Snow, C.D. | | Deposit date: | 2021-11-01 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Modular Protein-DNA Cocrystals as Precise, Programmable Assembly Scaffolds.
Acs Nano, 17, 2023
|
|
7SPM
 
 | | Replication Initiator Protein REPE54 and cognate DNA sequence with terminal three prime phosphates chemically crosslinked (30 mg/mL EDC, 12 hours, 2 doses). | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*A)-3'), DNA (5'-D(*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*A)-3'), MAGNESIUM ION, ... | | Authors: | Ward, A.R, Snow, C.D. | | Deposit date: | 2021-11-02 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Stabilizing DNA-Protein Co-Crystals via Intra-Crystal Chemical Ligation of the DNA
Crystals, 12, 2022
|
|
7SBG
 
 | |
6P0J
 
 | | Crystal structure of GDP-bound human RalA | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Ral-A | | Authors: | Bum-Erdene, K, Gonzalez-Gutierrez, G, Liu, D, Meroueh, S.O. | | Deposit date: | 2019-05-17 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Small-molecule covalent bond formation at tyrosine creates a binding site and inhibits activation of Ral GTPases.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PHJ
 
 | |
6PHO
 
 | |
6PHK
 
 | |
6PHP
 
 | |
6PHN
 
 | |
6PHM
 
 | |
6P7M
 
 | | Cryo-EM structure of LbCas12a-crRNA: AcrVA4 (1:2 complex) | | Descriptor: | Cas12a, MAGNESIUM ION, anti-CRISPR VA4, ... | | Authors: | Knott, G.J, Liu, J.J, Doudna, J.A. | | Deposit date: | 2019-06-06 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for AcrVA4 inhibition of specific CRISPR-Cas12a.
Elife, 8, 2019
|
|
6PHL
 
 | |
6PHQ
 
 | |
