6B2Q
 
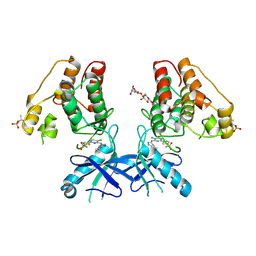 | | Dual Inhibition of the Essential Protein Kinases A and B in Mycobacterium tuberculosis | | Descriptor: | 3-methyl-1-(2-methylpropyl)butyl 4-O-beta-L-gulopyranosyl-beta-D-glucopyranoside, 5-{5-chloro-4-[(5-cyclopropyl-1H-pyrazol-3-yl)amino]pyrimidin-2-yl}thiophene-2-sulfonamide, Serine/threonine-protein kinase PknA | | Authors: | Zuccola, H.J. | | Deposit date: | 2017-09-20 | | Release date: | 2018-02-14 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Mtb PKNA/PKNB Dual Inhibition Provides Selectivity Advantages for Inhibitor Design To Minimize Host Kinase Interactions.
ACS Med Chem Lett, 8, 2017
|
|
6B2P
 
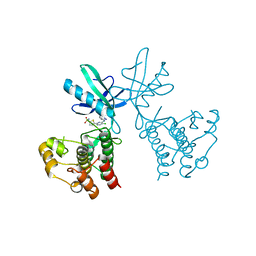 | | Dual Inhibition of the Essential Protein Kinases A and B in Mycobacterium tuberculosis | | Descriptor: | 5-{5-chloro-4-[(5-cyclopropyl-1H-pyrazol-3-yl)amino]pyrimidin-2-yl}thiophene-2-sulfonamide, Serine/threonine-protein kinase PknB | | Authors: | Zuccola, H.J. | | Deposit date: | 2017-09-20 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Mtb PKNA/PKNB Dual Inhibition Provides Selectivity Advantages for Inhibitor Design To Minimize Host Kinase Interactions.
ACS Med Chem Lett, 8, 2017
|
|
3I60
 
 | |
3I5Z
 
 | |
3I4B
 
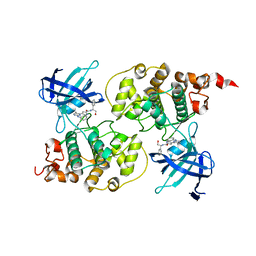 | | Crystal structure of GSK3b in complex with a pyrimidylpyrrole inhibitor | | Descriptor: | Glycogen synthase kinase-3 beta, N-[(1S)-2-hydroxy-1-phenylethyl]-4-[5-methyl-2-(phenylamino)pyrimidin-4-yl]-1H-pyrrole-2-carboxamide | | Authors: | Ter Haar, E. | | Deposit date: | 2009-07-01 | | Release date: | 2010-01-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided design of potent and selective pyrimidylpyrrole inhibitors of extracellular signal-regulated kinase (ERK) using conformational control.
J.Med.Chem., 52, 2009
|
|
2OJG
 
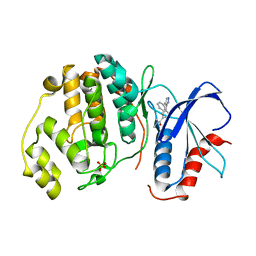 | |
2OJJ
 
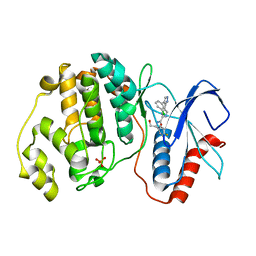 | |
2OJI
 
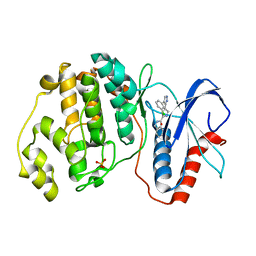 | |
2OK1
 
 | |
8QTO
 
 | | CRYSTAL STRUCTURE OF HOLO-L28H-FNR OF A. FISCHERI | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FNR type regulator, IRON/SULFUR CLUSTER | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2023-10-13 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Probing the Reactivity of [4Fe-4S] Fumarate and Nitrate Reduction (FNR) Regulator with O2 and NO: Increased O2 Resistance and Relative Specificity for NO of the [4Fe-4S] L28H FNR Cluster
Inorganics (Basel), 11, 2023
|
|
3G9L
 
 | |
3G9N
 
 | |
3G90
 
 | |
4PHV
 
 | | X-RAY CRYSTAL STRUCTURE OF THE HIV PROTEASE COMPLEX WITH L-700,417, AN INHIBITOR WITH PSEUDO C2 SYMMETRY | | Descriptor: | HIV-1 PROTEASE, N,N-BIS(2-HYDROXY-1-INDANYL)-2,6- DIPHENYLMETHYL-4-HYDROXY-1,7-HEPTANDIAMIDE | | Authors: | Bone, R. | | Deposit date: | 1991-10-04 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-Ray Crystal Structure of the HIV Protease Complex with L-700,417, an Inhibitor with Pseudo C2 Symmetry
J.Am.Chem.Soc., 113, 1991
|
|
3CYO
 
 | |
3CP1
 
 | |
1UBI
 
 | | SYNTHETIC STRUCTURAL AND BIOLOGICAL STUDIES OF THE UBIQUITIN SYSTEM. PART 1 | | Descriptor: | UBIQUITIN | | Authors: | Alexeev, D, Bury, S.M, Turner, M.A, Ogunjobi, O.M, Muir, T.W, Ramage, R, Sawyer, L. | | Deposit date: | 1994-02-03 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthetic, structural and biological studies of the ubiquitin system: the total chemical synthesis of ubiquitin.
Biochem.J., 299, 1994
|
|
1CR6
 
 | | CRYSTAL STRUCTURE OF MURINE SOLUBLE EPOXIDE HYDROLASE COMPLEXED WITH CPU INHIBITOR | | Descriptor: | EPOXIDE HYDROLASE, N-CYCLOHEXYL-N'-(PROPYL)PHENYL UREA | | Authors: | Argiriadi, M.A, Morisseau, C, Hammock, B.D, Christianson, D.W. | | Deposit date: | 1999-08-13 | | Release date: | 1999-11-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Detoxification of environmental mutagens and carcinogens: structure, mechanism, and evolution of liver epoxide hydrolase.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
