4U5E
 
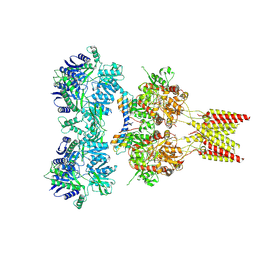 | | Crystal structure of GluA2 T625G, con-ikot-ikot snail toxin, partial agonist KA and postitive modulator (R,R)-2b complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Con-ikot-ikot, ... | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5073 Å) | | Cite: | X-ray structures of AMPA receptor-cone snail toxin complexes illuminate activation mechanism.
Science, 345, 2014
|
|
3F3A
 
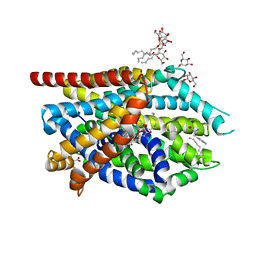 | | Crystal Structure of LeuT bound to L-Tryptophan and Sodium | | Descriptor: | SODIUM ION, TETRADECANE, TRYPTOPHAN, ... | | Authors: | Singh, S.K, Piscitelli, C.L, Yamashita, A, Gouaux, E. | | Deposit date: | 2008-10-30 | | Release date: | 2008-12-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A competitive inhibitor traps LeuT in an open-to-out conformation.
Science, 322, 2008
|
|
3F3D
 
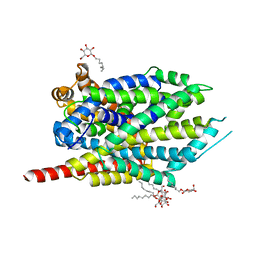 | | Crystal structure of LeuT bound to L-Methionine and sodium | | Descriptor: | METHIONINE, SODIUM ION, Transporter, ... | | Authors: | Singh, S.K, Piscitelli, C.L, Yamashita, A, Gouaux, E. | | Deposit date: | 2008-10-30 | | Release date: | 2008-12-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A competitive inhibitor traps LeuT in an open-to-out conformation.
Science, 322, 2008
|
|
3F3E
 
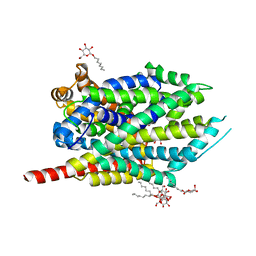 | | Crystal structure of LeuT bound to L-leucine (30 mM) and sodium | | Descriptor: | LEUCINE, SODIUM ION, Transporter, ... | | Authors: | Singh, S.K, Piscitelli, C.L, Yamashita, A, Gouaux, E. | | Deposit date: | 2008-10-30 | | Release date: | 2008-12-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A competitive inhibitor traps LeuT in an open-to-out conformation.
Science, 322, 2008
|
|
3F3C
 
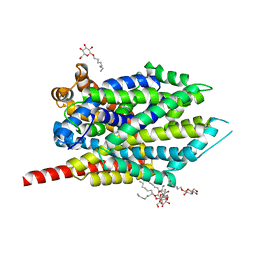 | | Crystal structure of LeuT bound to 4-Fluoro-L-Phenylalanine and sodium | | Descriptor: | 4-FLUORO-L-PHENYLALANINE, SODIUM ION, Transporter, ... | | Authors: | Singh, S.K, Piscitelli, C.L, Yamashita, A, Gouaux, E. | | Deposit date: | 2008-10-30 | | Release date: | 2008-12-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A competitive inhibitor traps LeuT in an open-to-out conformation.
Science, 322, 2008
|
|
3GIA
 
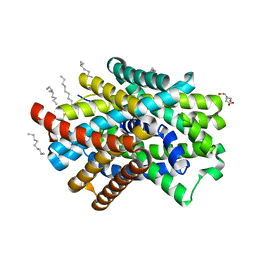 | | Crystal Structure of ApcT Transporter | | Descriptor: | BICINE, DECANE, Uncharacterized protein MJ0609 | | Authors: | Shaffer, P.L, Goehring, A.S, Shankaranarayanan, A, Gouaux, E, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2009-03-05 | | Release date: | 2009-08-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure and mechanism of a na+-independent amino Acid transporter.
Science, 325, 2009
|
|
3GI9
 
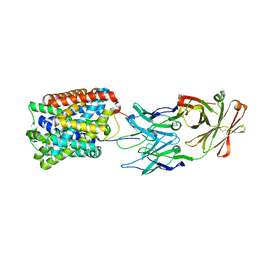 | | Crystal Structure of ApcT Transporter Bound to 7F11 Monoclonal Fab Fragment | | Descriptor: | 7F11 Anti-ApcT Monoclonal Fab Heavy Chain, 7F11 Anti-ApcT Monoclonal Fab Light Chain, Uncharacterized protein MJ0609 | | Authors: | Shaffer, P.L, Goehring, A.S, Shankaranarayanan, A, Gouaux, E, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2009-03-05 | | Release date: | 2009-08-18 | | Last modified: | 2017-06-07 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure and mechanism of a na+-independent amino Acid transporter.
Science, 325, 2009
|
|
3QS5
 
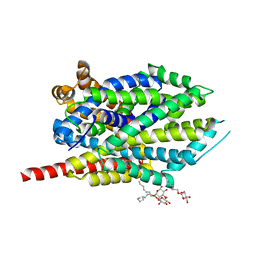 | |
3RIA
 
 | | C. elegans glutamate-gated chloride channel (GluCl) in complex with Fab, ivermectin and iodide. | | Descriptor: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, Avermectin-sensitive glutamate-gated chloride channel GluCl alpha, CHLORIDE ION, ... | | Authors: | Hibbs, R.E, Gouaux, E. | | Deposit date: | 2011-04-13 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Principles of activation and permeation in an anion-selective Cys-loop receptor.
Nature, 474, 2011
|
|
3RHW
 
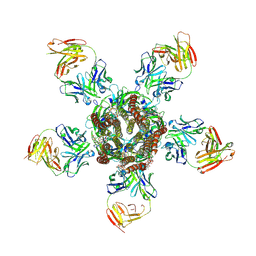 | | C. elegans glutamate-gated chloride channel (GluCl) in complex with Fab and ivermectin | | Descriptor: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, Avermectin-sensitive glutamate-gated chloride channel GluCl alpha, ... | | Authors: | Hibbs, R.E, Gouaux, E. | | Deposit date: | 2011-04-12 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | Principles of activation and permeation in an anion-selective Cys-loop receptor.
Nature, 474, 2011
|
|
3RI5
 
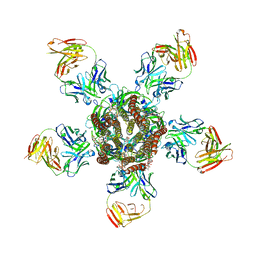 | | C. elegans glutamate-gated chloride channel (GluCl) in complex with Fab, ivermectin and picrotoxin | | Descriptor: | (1aR,2aR,3S,6R,6aS,8aS,8bR,9R)-2a-hydroxy-8b-methyl-9-(prop-1-en-2-yl)hexahydro-3,6-methano-1,5,7-trioxacyclopenta[ij]c yclopropa[a]azulene-4,8(3H)-dione, (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hibbs, R.E, Gouaux, E. | | Deposit date: | 2011-04-12 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Principles of activation and permeation in an anion-selective Cys-loop receptor.
Nature, 474, 2011
|
|
2A5T
 
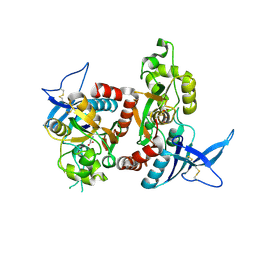 | | Crystal Structure Of The NR1/NR2A ligand-binding cores complex | | Descriptor: | GLUTAMIC ACID, GLYCINE, N-methyl-D-aspartate receptor NMDAR1-4a subunit, ... | | Authors: | Furukawa, H, Singh, S.K, Mancusso, R, Gouaux, E. | | Deposit date: | 2005-06-30 | | Release date: | 2005-11-15 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Subunit arrangement and function in NMDA receptors
Nature, 438, 2005
|
|
2A5S
 
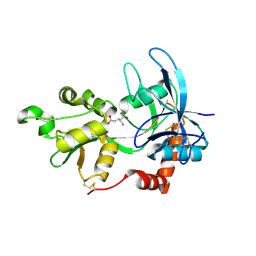 | | Crystal Structure Of The NR2A Ligand Binding Core In Complex With Glutamate | | Descriptor: | GLUTAMIC ACID, N-methyl-D-aspartate receptor NMDAR2A subunit | | Authors: | Furukawa, H, Singh, S.K, Mancusso, R, Gouaux, E. | | Deposit date: | 2005-06-30 | | Release date: | 2005-11-15 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Subunit arrangement and function in NMDA receptors
Nature, 438, 2005
|
|
3RIF
 
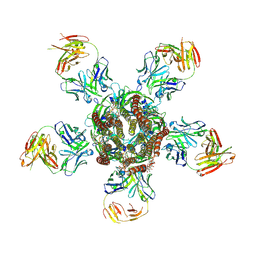 | | C. elegans glutamate-gated chloride channel (GluCl) in complex with Fab, ivermectin and glutamate. | | Descriptor: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, Avermectin-sensitive glutamate-gated chloride channel GluCl alpha, ... | | Authors: | Hibbs, R.E, Gouaux, E. | | Deposit date: | 2011-04-13 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.345 Å) | | Cite: | Principles of activation and permeation in an anion-selective Cys-loop receptor.
Nature, 474, 2011
|
|
3QS6
 
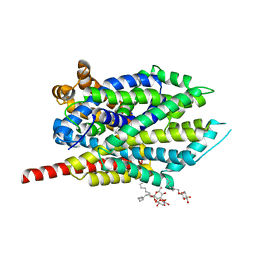 | |
3QS4
 
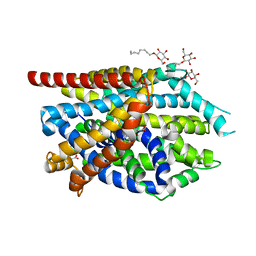 | |
5UN1
 
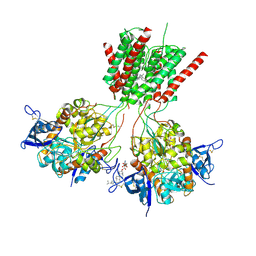 | | Crystal structure of GluN1/GluN2B delta-ATD NMDA receptor | | Descriptor: | (5S,10R)-5-methyl-10,11-dihydro-5H-5,10-epiminodibenzo[a,d][7]annulene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Song, X, Gouaux, E. | | Deposit date: | 2017-01-30 | | Release date: | 2018-02-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Mechanism of NMDA receptor channel block by MK-801 and memantine.
Nature, 556, 2018
|
|
5UP2
 
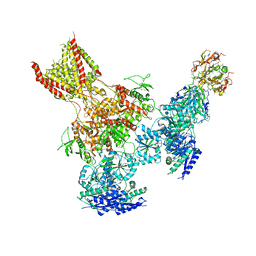 | | Triheteromeric NMDA receptor GluN1/GluN2A/GluN2B in complex with glycine, glutamate, Ro 25-6981, MK-801 and a GluN2B-specific Fab, at pH 6.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GluN2B-specific Fab, ... | | Authors: | Lu, W, Du, J, Goehring, A, Gouaux, E. | | Deposit date: | 2017-02-01 | | Release date: | 2017-03-22 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Cryo-EM structures of the triheteromeric NMDA receptor and its allosteric modulation.
Science, 355, 2017
|
|
5UOW
 
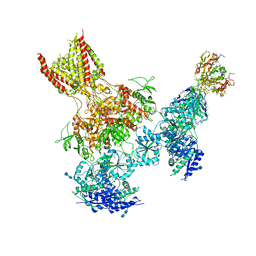 | | Triheteromeric NMDA receptor GluN1/GluN2A/GluN2B in complex with glycine, glutamate, MK-801 and a GluN2B-specific Fab, at pH 6.5 | | Descriptor: | (5S,10R)-5-methyl-10,11-dihydro-5H-5,10-epiminodibenzo[a,d][7]annulene, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Lu, W, Du, J, Goehring, A, Gouaux, E. | | Deposit date: | 2017-02-01 | | Release date: | 2017-03-22 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structures of the triheteromeric NMDA receptor and its allosteric modulation.
Science, 355, 2017
|
|
2A65
 
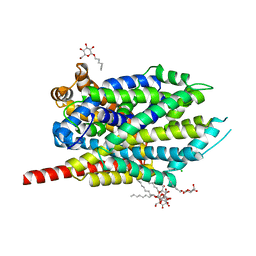 | | Crystal structure of LEUTAA, a bacterial homolog of Na+/Cl--dependent neurotransmitter transporters | | Descriptor: | CHLORIDE ION, LEUCINE, Na(+):neurotransmitter symporter (Snf family), ... | | Authors: | Yamashita, A, Singh, S.K, Kawate, T, Jin, Y, Gouaux, E. | | Deposit date: | 2005-07-01 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a bacterial homologue of Na(+)/Cl(-)-dependent neurotransmitter transporters.
Nature, 437, 2005
|
|
1LB8
 
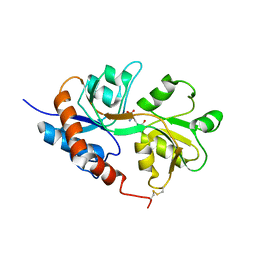 | | Crystal structure of the Non-desensitizing GluR2 ligand binding core mutant (S1S2J-L483Y) in complex with AMPA at 2.3 resolution | | Descriptor: | (S)-ALPHA-AMINO-3-HYDROXY-5-METHYL-4-ISOXAZOLEPROPIONIC ACID, Glutamate receptor 2 | | Authors: | Sun, Y, Olson, R, Horning, M, Armstrong, N, Mayer, M, Gouaux, E. | | Deposit date: | 2002-04-02 | | Release date: | 2002-06-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of glutamate receptor desensitization.
Nature, 417, 2002
|
|
1LB9
 
 | | Crystal structure of the Non-desensitizing GluR2 ligand binding core mutant (S1S2J-L483Y) in complex with antagonist DNQX at 2.3 A resolution | | Descriptor: | 6,7-DINITROQUINOXALINE-2,3-DIONE, Glutamate receptor 2, SULFATE ION | | Authors: | Sun, Y, Olson, R, Horning, M, Armstrong, N, Mayer, M, Gouaux, E. | | Deposit date: | 2002-04-02 | | Release date: | 2002-06-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of glutamate receptor desensitization.
Nature, 417, 2002
|
|
1LBB
 
 | | Crystal structure of the GluR2 ligand binding domain mutant (S1S2J-N754D) in complex with kainate at 2.1 A resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamine receptor 2 | | Authors: | Sun, Y, Olson, R, Horning, M, Armstrong, N, Mayer, M, Gouaux, E. | | Deposit date: | 2002-04-02 | | Release date: | 2002-06-05 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of glutamate receptor desensitization.
Nature, 417, 2002
|
|
4XP4
 
 | | X-ray structure of Drosophila dopamine transporter in complex with cocaine | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, ANTIBODY FRAGMENT HEAVY CHAIN-PROTEIN, 9D5-HEAVY CHAIN, ... | | Authors: | Aravind, P, Wang, K, Gouaux, E. | | Deposit date: | 2015-01-16 | | Release date: | 2015-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Neurotransmitter and psychostimulant recognition by the dopamine transporter.
Nature, 521, 2015
|
|
4XNU
 
 | | X-ray structure of Drosophila dopamine transporter in complex with nisoxetine | | Descriptor: | CHLORIDE ION, CHOLESTEROL, Integral membrane protein-dopamine transporter, ... | | Authors: | Aravind, P, Wang, K, Gouaux, E. | | Deposit date: | 2015-01-16 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | X-ray structures of Drosophila dopamine transporter in complex with nisoxetine and reboxetine.
Nat.Struct.Mol.Biol., 22, 2015
|
|
