7E7Q
 
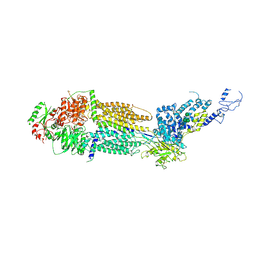 | | Cryo-EM structure of human ABCA4 in ATP-bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xie, T, Zhang, Z.K, Gong, X. | | Deposit date: | 2021-02-26 | | Release date: | 2021-06-30 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of substrate recognition and translocation by human ABCA4.
Nat Commun, 12, 2021
|
|
7E7O
 
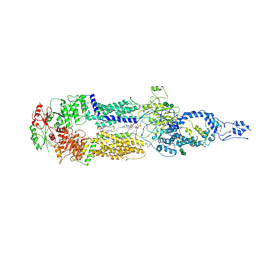 | | Cryo-EM structure of human ABCA4 in NRPE-bound state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Retinal-specific phospholipid-transporting ATPase ABCA4, ... | | Authors: | Xie, T, Zhang, Z.K, Gong, X. | | Deposit date: | 2021-02-26 | | Release date: | 2021-06-30 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of substrate recognition and translocation by human ABCA4.
Nat Commun, 12, 2021
|
|
7E2G
 
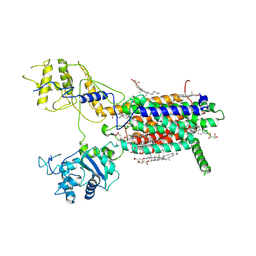 | | Cryo-EM structure of hDisp1NNN-3C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Protein dispatched homolog 1,Protein dispatched homolog 1 | | Authors: | Li, W, Wang, L, Gong, X. | | Deposit date: | 2021-02-05 | | Release date: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Structural insights into proteolytic activation of the human Dispatched1 transporter for Hedgehog morphogen release.
Nat Commun, 12, 2021
|
|
7E2I
 
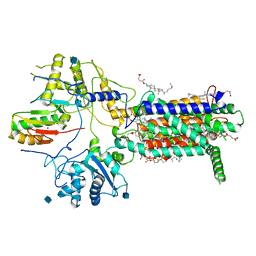 | | Cryo-EM structure of hDisp1NNN-ShhN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Protein dispatched homolog 1, ... | | Authors: | Li, W, Wang, L, Gong, X. | | Deposit date: | 2021-02-05 | | Release date: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Structural insights into proteolytic activation of the human Dispatched1 transporter for Hedgehog morphogen release.
Nat Commun, 12, 2021
|
|
7E2H
 
 | | Cryo-EM structure of hDisp1NNN-3C-Cleavage | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Protein dispatched homolog 1 | | Authors: | Li, W, Wang, L, Gong, X. | | Deposit date: | 2021-02-05 | | Release date: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structural insights into proteolytic activation of the human Dispatched1 transporter for Hedgehog morphogen release.
Nat Commun, 12, 2021
|
|
4TTQ
 
 | |
4IA0
 
 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 5-bromo-2-{2-ethoxy-5-[(4-methylpiperazin-1-yl)sulfonyl]phenyl}-6-octylpyrimidin-4(3H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Ren, J, Chen, T, Xu, Y. | | Deposit date: | 2012-12-05 | | Release date: | 2014-01-01 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Exploration of the 5-bromopyrimidin-4(3H)-ones as potent inhibitors of PDE5.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4I9Z
 
 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 5-bromo-2-{5-[(4-methylpiperazin-1-yl)acetyl]-2-propoxyphenyl}-6-(propan-2-yl)pyrimidin-4(3H)-one, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Ren, J, Chen, T, Xu, Y. | | Deposit date: | 2012-12-05 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Exploration of the 5-bromopyrimidin-4(3H)-ones as potent inhibitors of PDE5.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4TTR
 
 | |
6C2R
 
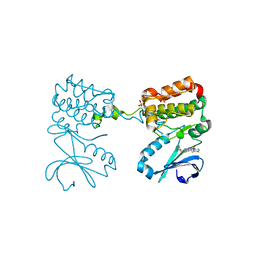 | | Aurora A ligand complex | | Descriptor: | (2R,4R)-1-[(3-chloro-2-fluorophenyl)methyl]-4-({3-fluoro-6-[(5-methyl-1H-pyrazol-3-yl)amino]pyridin-2-yl}methyl)-2-methylpiperidine-4-carboxylic acid, Aurora kinase A, SULFATE ION | | Authors: | Antonysamy, S, Pustilnik, A, Manglicmot, D, Froning, K, Weichert, K, Wasserman, S. | | Deposit date: | 2018-01-08 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Aurora A Kinase Inhibition Is Synthetic Lethal with Loss of theRB1Tumor Suppressor Gene.
Cancer Discov, 9, 2019
|
|
6C2T
 
 | | Aurora A ligand complex | | Descriptor: | (2S,4R)-1-[(3-chloro-2-fluorophenyl)methyl]-2-methyl-4-({3-[(1,3-thiazol-2-yl)amino]isoquinolin-1-yl}methyl)piperidine-4-carboxylic acid, Aurora kinase A, DIMETHYL SULFOXIDE, ... | | Authors: | Antonysamy, S, Pustilnik, A, Manglicmot, D, Froning, K, Weichert, K, Wasserman, S. | | Deposit date: | 2018-01-08 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Aurora A Kinase Inhibition Is Synthetic Lethal with Loss of theRB1Tumor Suppressor Gene.
Cancer Discov, 9, 2019
|
|
7F69
 
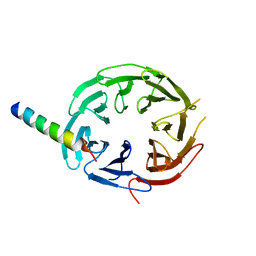 | | Crystal structure of WIPI2b in complex with ATG16L1 | | Descriptor: | Isoform 2 of Autophagy-related protein 16-1, Isoform 2 of WD repeat domain phosphoinositide-interacting protein 2 | | Authors: | Gong, X.Y, Pan, L.F. | | Deposit date: | 2021-06-24 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | ATG16L1 adopts a dual-binding site mode to interact with WIPI2b in autophagy.
Sci Adv, 9, 2023
|
|
7XFR
 
 | |
4XU5
 
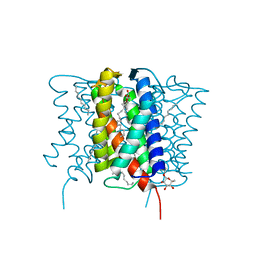 | | Crystal structure of MvINS bound to a bromine-derived 14C Diacylglycerol (DAG) at 2.1A resolution | | Descriptor: | (2S)-1-[(13-bromotridecanoyl)oxy]-3-hydroxypropan-2-yl tetradecanoate, DECANE, Uncharacterized protein, ... | | Authors: | Ren, R.B, Wu, J.P, Yan, C.Y, He, Y, Yan, N. | | Deposit date: | 2015-01-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | PROTEIN STRUCTURE. Crystal structure of a mycobacterial Insig homolog provides insight into how these sensors monitor sterol levels
Science, 349, 2015
|
|
4XU4
 
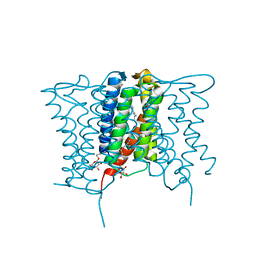 | | Crystal structure of a mycobacterial Insig homolog MvINS from Mycobacterium vanbaalenii at 1.9A resolution | | Descriptor: | DECYLAMINE-N,N-DIMETHYL-N-OXIDE, Uncharacterized protein, nonyl beta-D-glucopyranoside | | Authors: | Ren, R.B, Wu, J.P, Yan, C.Y, He, Y, Yan, N. | | Deposit date: | 2015-01-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | PROTEIN STRUCTURE. Crystal structure of a mycobacterial Insig homolog provides insight into how these sensors monitor sterol levels
Science, 349, 2015
|
|
4XU6
 
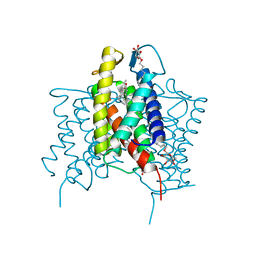 | | Crystal structure of cross-linked MvINS R77C trimer at 1.9A resolution | | Descriptor: | N-TRIDECANOIC ACID, Uncharacterized protein, octyl beta-D-glucopyranoside | | Authors: | Ren, R.B, Wu, J.P, Yan, C.Y, He, Y, Yan, N. | | Deposit date: | 2015-01-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | PROTEIN STRUCTURE. Crystal structure of a mycobacterial Insig homolog provides insight into how these sensors monitor sterol levels
Science, 349, 2015
|
|
7YUJ
 
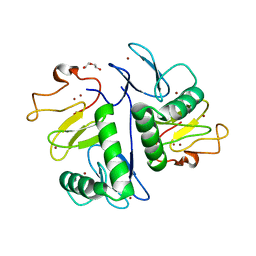 | | Crystal structure of HOIL-1L(365-510) | | Descriptor: | DI(HYDROXYETHYL)ETHER, RanBP-type and C3HC4-type zinc finger-containing protein 1, ZINC ION | | Authors: | Xiao, L, Pan, L. | | Deposit date: | 2022-08-17 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.865 Å) | | Cite: | Mechanistic insights into the enzymatic activity of E3 ligase HOIL-1L and its regulation by the linear ubiquitin chain binding.
Sci Adv, 9, 2023
|
|
7YUI
 
 | |
5C9C
 
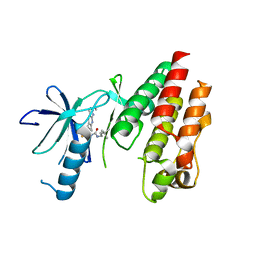 | | CRYSTAL STRUCTURE OF BRAF(V600E) IN COMPLEX WITH LY3009120 COMPND | | Descriptor: | 1-(3,3-dimethylbutyl)-3-{2-fluoro-4-methyl-5-[7-methyl-2-(methylamino)pyrido[2,3-d]pyrimidin-6-yl]phenyl}urea, CHLORIDE ION, Serine/threonine-protein kinase B-raf | | Authors: | Edwards, T, Abendroth, J, Chun, L. | | Deposit date: | 2015-06-26 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Inhibition of RAF Isoforms and Active Dimers by LY3009120 Leads to Anti-tumor Activities in RAS or BRAF Mutant Cancers.
Cancer Cell, 28, 2015
|
|
6IWI
 
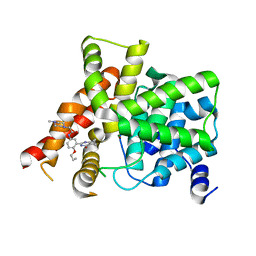 | | Crystal structure of PDE5A in complex with a novel inhibitor | | Descriptor: | MAGNESIUM ION, N-[3-(4,5-diethyl-6-oxo-1,6-dihydropyrimidin-2-yl)-4-propoxyphenyl]-2-(4-methylpiperazin-1-yl)acetamide, ZINC ION, ... | | Authors: | Zhang, X.L, Xu, Y.C. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.155 Å) | | Cite: | Pharmacokinetics-Driven Optimization of 4(3 H)-Pyrimidinones as Phosphodiesterase Type 5 Inhibitors Leading to TPN171, a Clinical Candidate for the Treatment of Pulmonary Arterial Hypertension.
J.Med.Chem., 62, 2019
|
|
4GBY
 
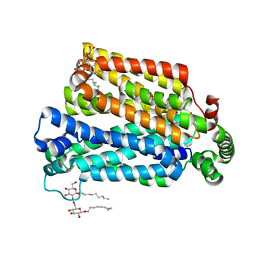 | | The structure of the MFS (major facilitator superfamily) proton:xylose symporter XylE bound to D-xylose | | Descriptor: | D-xylose-proton symporter, beta-D-xylopyranose, nonyl beta-D-glucopyranoside | | Authors: | Sun, L.F, Zeng, X, Yan, C.Y, Yan, N. | | Deposit date: | 2012-07-28 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.808 Å) | | Cite: | Crystal structure of a bacterial homologue of glucose transporters GLUT1-4.
Nature, 490, 2012
|
|
4GBZ
 
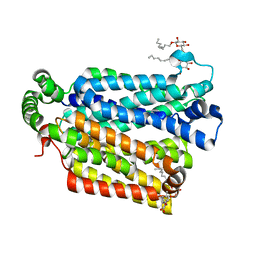 | | The structure of the MFS (major facilitator superfamily) proton:xylose symporter XylE bound to D-glucose | | Descriptor: | D-xylose-proton symporter, beta-D-glucopyranose, nonyl beta-D-glucopyranoside | | Authors: | Sun, L.F, Zeng, X, Yan, C.Y, Yan, N. | | Deposit date: | 2012-07-28 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Crystal structure of a bacterial homologue of glucose transporters GLUT1-4.
Nature, 490, 2012
|
|
4GC0
 
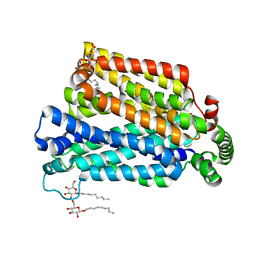 | | The structure of the MFS (major facilitator superfamily) proton:xylose symporter XylE bound to 6-bromo-6-deoxy-D-glucose | | Descriptor: | 6-bromo-6-deoxy-beta-D-glucopyranose, D-xylose-proton symporter, nonyl beta-D-glucopyranoside | | Authors: | Yan, N, Sun, L.F, Zeng, X, Yan, C.Y. | | Deposit date: | 2012-07-28 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a bacterial homologue of glucose transporters GLUT1-4.
Nature, 490, 2012
|
|
6IWW
 
 | | Cryo-EM structure of the S. typhimurium oxaloacetate decarboxylase beta-gamma sub-complex | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Oxaloacetate decarboxylase beta chain, Probable oxaloacetate decarboxylase gamma chain | | Authors: | Xu, X, Shi, H, Zhang, X, Xiang, S. | | Deposit date: | 2018-12-08 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into sodium transport by the oxaloacetate decarboxylase sodium pump.
Elife, 9, 2020
|
|
6IVA
 
 | |
