3OL6
 
 | | Poliovirus polymerase elongation complex | | Descriptor: | ISOPROPYL ALCOHOL, Polymerase, RNA (5'-R(*AP*AP*GP*UP*CP*UP*CP*CP*AP*GP*GP*UP*CP*UP*CP*UP*CP*GP*UP*CP*CP*GP*GP*AP*AP*A)-3'), ... | | Authors: | Gong, P, Peersen, O.B. | | Deposit date: | 2010-08-25 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OLA
 
 | | Poliovirus polymerase elongation complex with 2'-deoxy-CTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, DNA/RNA (5'-R(*GP*CP*CP*CP*GP*GP*AP*CP*GP*AP*GP*AP*GP*A)-D(P*C)-3'), ... | | Authors: | Gong, P, Peersen, O.B. | | Deposit date: | 2010-08-25 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OL9
 
 | |
3OL7
 
 | | Poliovirus polymerase elongation complex with CTP | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Gong, P, Peersen, O.B. | | Deposit date: | 2010-08-25 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OL8
 
 | | Poliovirus polymerase elongation complex with CTP-Mn | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, MANGANESE (II) ION, ... | | Authors: | Gong, P, Peersen, O.B. | | Deposit date: | 2010-08-25 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OLB
 
 | | Poliovirus polymerase elongation complex with 2',3'-dideoxy-ctp | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, ISOPROPYL ALCOHOL, Polymerase, ... | | Authors: | Gong, P, Peersen, O.B. | | Deposit date: | 2010-08-25 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4K4Y
 
 | | Coxsackievirus B3 polymerase elongation complex (r2+1_form) | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, ACETATE ION, DNA/RNA (5'-R(*UP*GP*UP*UP*CP*GP*AP*CP*GP*AP*GP*AP*GP*AP*(DOC))-3'), ... | | Authors: | Gong, P, Peersen, O.B. | | Deposit date: | 2013-04-12 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structures of coxsackievirus, rhinovirus, and poliovirus polymerase elongation complexes solved by engineering RNA mediated crystal contacts.
Plos One, 8, 2013
|
|
4K4T
 
 | | Poliovirus polymerase elongation complex (r4_form) | | Descriptor: | GLYCEROL, RNA (5'-R(*AP*AP*GP*UP*CP*UP*CP*CP*AP*GP*GP*UP*CP*UP*CP*UP*CP*GP*GP*AP*AP*A)-3'), RNA (5'-R(*GP*GP*GP*AP*GP*AP*UP*GP*A)-3'), ... | | Authors: | Gong, P, Peersen, O.B. | | Deposit date: | 2013-04-12 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structures of coxsackievirus, rhinovirus, and poliovirus polymerase elongation complexes solved by engineering RNA mediated crystal contacts.
Plos One, 8, 2013
|
|
4K4Z
 
 | | Coxsackievirus B3 polymerase elongation complex (r2_Mg_form) | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*AP*AP*GP*UP*CP*UP*CP*CP*AP*GP*GP*UP*CP*UP*CP*UP*CP*GP*UP*CP*GP*AP*AP*A)-3'), RNA (5'-R(*UP*GP*UP*UP*CP*GP*AP*CP*GP*AP*GP*AP*GP*A)-3'), ... | | Authors: | Gong, P, Peersen, O.B. | | Deposit date: | 2013-04-12 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structures of coxsackievirus, rhinovirus, and poliovirus polymerase elongation complexes solved by engineering RNA mediated crystal contacts.
Plos One, 8, 2013
|
|
4K4U
 
 | | Poliovirus polymerase elongation complex (r5_form) | | Descriptor: | RNA (5'-R(*AP*AP*GP*UP*CP*UP*CP*CP*AP*GP*GP*UP*CP*UP*CP*UP*CP*UP*CP*GP*UP*CP*GP*AP*AP*A)-3'), RNA (5'-R(*UP*GP*UP*UP*CP*GP*AP*CP*GP*AP*GP*AP*GP*AP*GP*A)-3'), RNA (5'-R(P*GP*GP*GP*GP*GP*AP*GP*AP*UP*GP*A)-3'), ... | | Authors: | Gong, P, Peersen, O.B. | | Deposit date: | 2013-04-12 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structures of coxsackievirus, rhinovirus, and poliovirus polymerase elongation complexes solved by engineering RNA mediated crystal contacts.
Plos One, 8, 2013
|
|
4K4V
 
 | | Poliovirus polymerase elongation complex (r5+1_form) | | Descriptor: | DNA/RNA (5'-R(*UP*GP*UP*UP*CP*GP*AP*CP*GP*AP*GP*AP*GP*AP*GP*A)-D(P*C)-3'), RNA (5'-R(*AP*AP*GP*UP*CP*UP*CP*CP*AP*GP*GP*UP*CP*UP*CP*UP*CP*UP*CP*GP*UP*CP*GP*AP*AP*A)-3'), RNA-directed RNA polymerase 3D-POL, ... | | Authors: | Gong, P, Peersen, O.B. | | Deposit date: | 2013-04-12 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structures of coxsackievirus, rhinovirus, and poliovirus polymerase elongation complexes solved by engineering RNA mediated crystal contacts.
Plos One, 8, 2013
|
|
4K4X
 
 | | Coxsackievirus B3 polymerase elongation complex (r2_form), rna | | Descriptor: | GLYCEROL, MAGNESIUM ION, RNA (5'-R(*AP*AP*GP*UP*CP*UP*CP*CP*AP*GP*GP*UP*CP*UP*CP*UP*CP*GP*UP*CP*GP*AP*AP*A)-3'), ... | | Authors: | Gong, P, Peersen, O.B. | | Deposit date: | 2013-04-12 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structures of coxsackievirus, rhinovirus, and poliovirus polymerase elongation complexes solved by engineering RNA mediated crystal contacts.
Plos One, 8, 2013
|
|
4K50
 
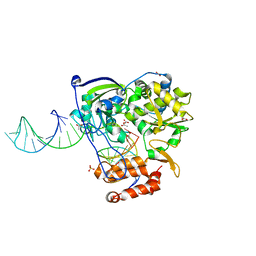 | | Rhinovirus 16 polymerase elongation complex (r1_form) | | Descriptor: | ACETATE ION, GLYCEROL, RNA (33-MER), ... | | Authors: | Gong, P, Peersen, O.B. | | Deposit date: | 2013-04-12 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structures of coxsackievirus, rhinovirus, and poliovirus polymerase elongation complexes solved by engineering RNA mediated crystal contacts.
Plos One, 8, 2013
|
|
4K4S
 
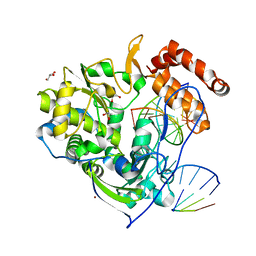 | | Poliovirus polymerase elongation complex (r3_form) | | Descriptor: | GLYCEROL, RNA (5'-R(*AP*AP*GP*UP*CP*UP*CP*CP*AP*GP*GP*UP*CP*UP*CP*UP*CP*GP*CP*GP*AP*AP*A)-3'), RNA (5'-R(*GP*GP*GP*AP*GP*AP*UP*GP*A)-3'), ... | | Authors: | Gong, P, Peersen, O.B. | | Deposit date: | 2013-04-12 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of coxsackievirus, rhinovirus, and poliovirus polymerase elongation complexes solved by engineering RNA mediated crystal contacts.
Plos One, 8, 2013
|
|
4K4W
 
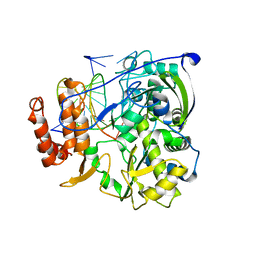 | | Poliovirus polymerase elongation complex (r5+2_form) | | Descriptor: | RNA (5'-R(*UP*GP*UP*UP*CP*GP*AP*CP*GP*AP*GP*AP*GP*AP*GP*AP*CP*C)-3'), RNA (5'-R(P*GP*GP*GP*AP*GP*AP*UP*GP*AP*AP*AP*GP*UP*CP*UP*CP*CP*AP*GP*GP*UP*CP*UP*CP*UP*CP*UP*CP*GP*UP*CP*GP*AP*AP*A)-3'), RNA-directed RNA polymerase 3D-POL | | Authors: | Gong, P, Peersen, O.B. | | Deposit date: | 2013-04-12 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structures of coxsackievirus, rhinovirus, and poliovirus polymerase elongation complexes solved by engineering RNA mediated crystal contacts.
Plos One, 8, 2013
|
|
8IIC
 
 | |
8IIB
 
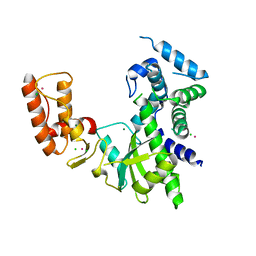 | | Crystal structure of Israeli acute paralysis virus RNA-dependent RNA polymerase delta85 mutant (residues 86-546) | | Descriptor: | CADMIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Fang, X, Lu, G, Hou, C, Gong, P. | | Deposit date: | 2023-02-24 | | Release date: | 2023-06-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Unusual substructure conformations observed in crystal structures of a dicistrovirus RNA-dependent RNA polymerase suggest contribution of the N-terminal extension in proper folding.
Virol Sin, 38, 2023
|
|
8I6G
 
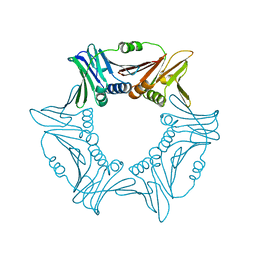 | |
8I6H
 
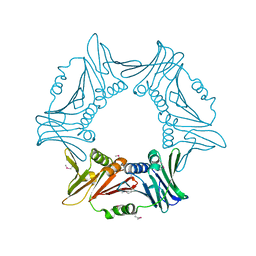 | |
6LSE
 
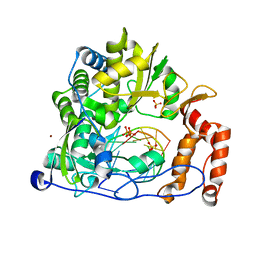 | | Crystal structure of the enterovirus 71 polymerase elongation complex (C3S6A/C3S6B form) | | Descriptor: | Genome polyprotein, PYROPHOSPHATE 2-, RNA (35-MER), ... | | Authors: | Wang, M, Shu, B, Jing, X, Gong, P. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Stringent control of the RNA-dependent RNA polymerase translocation revealed by multiple intermediate structures.
Nat Commun, 11, 2020
|
|
6LSF
 
 | | Crystal structure of the enterovirus 71 polymerase elongation complex (C2S6RA/C2S6RB form) | | Descriptor: | Genome polyprotein, RNA (35-MER), RNA (5'-R(*UP*GP*UP*UP*CP*GP*AP*CP*GP*AP*GP*AP*GP*AP*GP*AP*CP*C)-3'), ... | | Authors: | Wang, M, Shu, B, Jing, X, Gong, P. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Stringent control of the RNA-dependent RNA polymerase translocation revealed by multiple intermediate structures.
Nat Commun, 11, 2020
|
|
7D6N
 
 | |
7D6M
 
 | |
3HJH
 
 | | A rigid N-terminal clamp restrains the motor domains of the bacterial transcription-repair coupling factor | | Descriptor: | COBALT (II) ION, Transcription-repair-coupling factor | | Authors: | Murphy, M, Gong, P, Ralto, K, Manelyte, L, Savery, N, Theis, K. | | Deposit date: | 2009-05-21 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | An N-terminal clamp restrains the motor domains of the bacterial transcription-repair coupling factor Mfd.
Nucleic Acids Res., 37, 2009
|
|
6LSH
 
 | | Crystal structure of the enterovirus 71 polymerase elongation complex (C2S6M form) | | Descriptor: | Genome polyprotein, RNA (35-MER), RNA (5'-R(*UP*GP*UP*UP*CP*GP*AP*CP*GP*AP*GP*AP*GP*AP*GP*AP*CP*C)-3'), ... | | Authors: | Li, R, Wang, M, Jing, X, Gong, P. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | Stringent control of the RNA-dependent RNA polymerase translocation revealed by multiple intermediate structures.
Nat Commun, 11, 2020
|
|
