2UUR
 
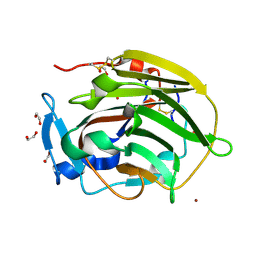 | | N-terminal NC4 domain of collagen IX | | Descriptor: | 1,2-ETHANEDIOL, COLLAGEN ALPHA-1(IX) CHAIN, SULFATE ION, ... | | Authors: | Leppanen, V.-M, Tossavainen, H, Permi, P, Lehtio, L, Ronnholm, G, Goldman, A, Kilpelainen, I, Pihlajamaa, T. | | Deposit date: | 2007-03-07 | | Release date: | 2007-06-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the N-Terminal Nc4 Domain of Collagen Ix, a Zinc Binding Member of the Laminin-Neurexin-Sex Hormone Binding Globulin (Lns) Domain Family.
J.Biol.Chem., 282, 2007
|
|
1QEZ
 
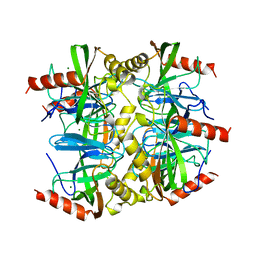 | | SULFOLOBUS ACIDOCALDARIUS INORGANIC PYROPHOSPHATASE: AN ARCHAEL PYROPHOSPHATASE. | | Descriptor: | MAGNESIUM ION, PROTEIN (INORGANIC PYROPHOSPHATASE) | | Authors: | Leppanen, V.-M, Nummelin, H, Hansen, T, Lahti, R, Schafer, G, Goldman, A. | | Deposit date: | 1999-04-06 | | Release date: | 1999-04-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Sulfolobus acidocaldarius inorganic pyrophosphatase: structure, thermostability, and effect of metal ion in an archael pyrophosphatase.
Protein Sci., 8, 1999
|
|
1YQ5
 
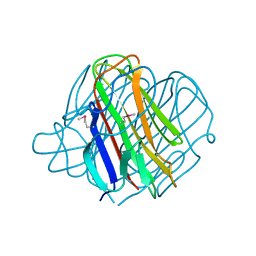 | | PRD1 vertex protein P5 | | Descriptor: | Minor capsid protein | | Authors: | Merckel, M.C, Huiskonen, J.T, Goldman, A, Bamford, D.H, Tuma, R. | | Deposit date: | 2005-02-01 | | Release date: | 2005-04-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the bacteriophage PRD1 spike sheds light on the evolution of viral capsid architecture.
Mol.Cell, 18, 2005
|
|
4KFU
 
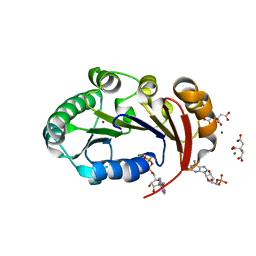 | | Structure of the genome packaging NTPase B204 from Sulfolobus turreted icosahedral virus 2 in complex with AMPPCP | | Descriptor: | CITRATE ANION, Genome packaging NTPase B204, MAGNESIUM ION, ... | | Authors: | Happonen, L.J, Oksanen, E, Kajander, T, Goldman, A, Butcher, S. | | Deposit date: | 2013-04-27 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | The Structure of the NTPase That Powers DNA Packaging into Sulfolobus Turreted Icosahedral Virus 2.
J.Virol., 87, 2013
|
|
2X1X
 
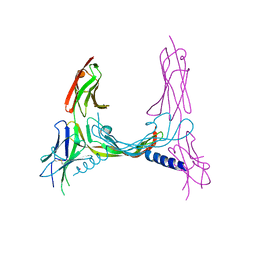 | | CRYSTAL STRUCTURE OF VEGF-C IN COMPLEX WITH DOMAINS 2 AND 3 OF VEGFR2 IN A TETRAGONAL CRYSTAL FORM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MERCURY (II) ION, ... | | Authors: | Leppanen, V.-M, Prota, A.E, Jeltsch, M, Anisimov, A, Kalkkinen, N, Strandin, T, Lankinen, H, Goldman, A, Ballmer-Hofer, K, Alitalo, K. | | Deposit date: | 2010-01-08 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Determinants of Growth Factor Binding and Specificity by Vegf Receptor 2.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2X1W
 
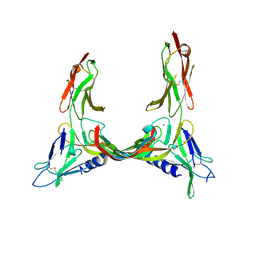 | | Crystal Structure of VEGF-C in Complex with Domains 2 and 3 of VEGFR2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CESIUM ION, ... | | Authors: | Leppanen, V.M, Prota, A.E, Jeltsch, M, Anisimov, A, Kalkkinen, N, Strandin, T, Lankinen, H, Goldman, A, Ballmer-Hofer, K, Alitalo, K. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Determinants of Growth Factor Binding and Specificity by Vegf Receptor 2.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2XQH
 
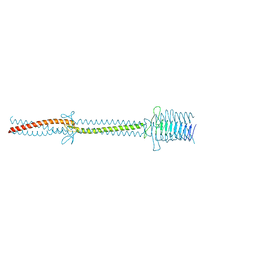 | | Crystal structure of an immunoglobulin-binding fragment of the trimeric autotransporter adhesin EibD | | Descriptor: | CHLORIDE ION, IMMUNOGLOBULIN-BINDING PROTEIN EIBD | | Authors: | Leo, J.C, Lyskowski, A, Hartmann, M, Schwarz, H, Linke, D, Lupas, A.N, Goldman, A. | | Deposit date: | 2010-09-02 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The Structure of E. Coli Igg-Binding Protein D Suggests a General Model for Bending and Binding in Trimeric Autotransporter Adhesins.
Structure, 19, 2011
|
|
4KFR
 
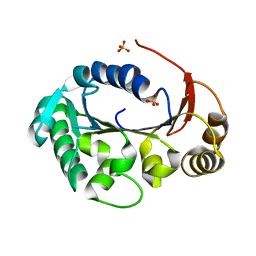 | | Structure of the genome packaging NTPase B204 from Sulfolobus turreted icosahedral virus 2 in complex with sulfate | | Descriptor: | Genome packaging NTPase B204, MAGNESIUM ION, SULFATE ION | | Authors: | Happonen, L.J, Oksanen, E, Goldman, A, Kajander, T, Butcher, S. | | Deposit date: | 2013-04-27 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.956 Å) | | Cite: | The Structure of the NTPase That Powers DNA Packaging into Sulfolobus Turreted Icosahedral Virus 2.
J.Virol., 87, 2013
|
|
4KFS
 
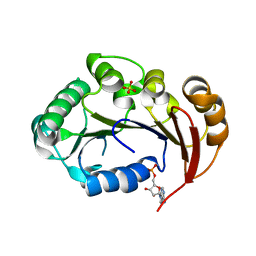 | | Structure of the genome packaging NTPase B204 from Sulfolobus turreted icosahedral virus 2 in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, CITRATE ANION, Genome packaging NTPase B204, ... | | Authors: | Happonen, L.J, Oksanen, E, Kajander, T, Goldman, A, Butcher, S. | | Deposit date: | 2013-04-27 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.946 Å) | | Cite: | The Structure of the NTPase That Powers DNA Packaging into Sulfolobus Turreted Icosahedral Virus 2.
J.Virol., 87, 2013
|
|
4KFT
 
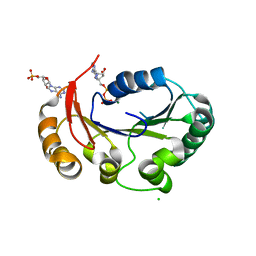 | | Structure of the genome packaging NTPase B204 from Sulfolobus turreted icosahedral virus 2 in complex with ATP-gammaS | | Descriptor: | CHLORIDE ION, CITRATE ANION, Genome packaging NTPase B204, ... | | Authors: | Happonen, L.J, Oksanen, E, Kajander, T, Goldman, A, Butcher, S. | | Deposit date: | 2013-04-27 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.241 Å) | | Cite: | The Structure of the NTPase That Powers DNA Packaging into Sulfolobus Turreted Icosahedral Virus 2.
J.Virol., 87, 2013
|
|
1MZO
 
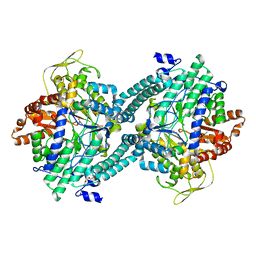 | | Crystal structure of pyruvate formate-lyase with pyruvate | | Descriptor: | PYRUVIC ACID, Pyruvate formate-lyase, TRIETHYLENE GLYCOL | | Authors: | Lehtio, L, Leppanen, V.-M, Kozarich, J.W, Goldman, A. | | Deposit date: | 2002-10-09 | | Release date: | 2002-12-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Escherichia coli pyruvate formate-lyase with pyruvate.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1NU5
 
 | |
1OIO
 
 | | GafD (F17c-type) Fimbrial adhesin from Escherichia coli | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FIMBRIAL LECTIN | | Authors: | Merckel, M.C, Tanskanen, J, Edelman, S, Westerlund-Wikstrom, B, Korhonen, T.K, Goldman, A. | | Deposit date: | 2003-06-22 | | Release date: | 2003-08-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structural Basis of Receptor-Binding by Escherichia Coli Associated with Diarrhea and Septicemia
J.Mol.Biol., 331, 2003
|
|
1P9H
 
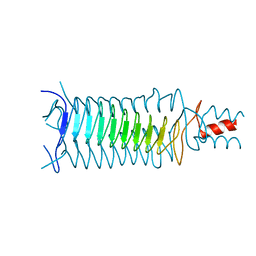 | |
2FEL
 
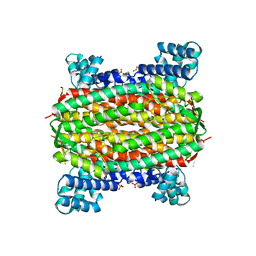 | | 3-carboxy-cis,cis-muconate lactonizing enzyme from Agrobacterium radiobacter S2 | | Descriptor: | 3-carboxy-cis,cis-muconate lactonizing enzyme, CHLORIDE ION, SULFATE ION | | Authors: | Lehtio, L, Goldman, A. | | Deposit date: | 2005-12-16 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of the 3-carboxy-cis,cis-muconate lactonizing enzyme from the protocatechuate degradative pathway of Agrobacterium radiobacter S2.
Febs J., 273, 2006
|
|
2FEN
 
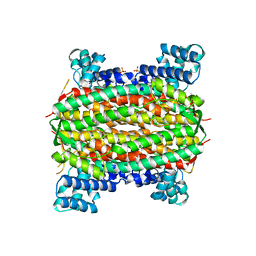 | | 3-carboxy-cis,cis-muconate lactonizing enzyme from Agrobacterium radiobacter S2 | | Descriptor: | 3-carboxy-cis,cis-muconate lactonizing enzyme, CHLORIDE ION, SULFATE ION | | Authors: | Lehtio, L, Goldman, A. | | Deposit date: | 2005-12-16 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and function of the 3-carboxy-cis,cis-muconate lactonizing enzyme from the protocatechuate degradative pathway of Agrobacterium radiobacter S2.
Febs J., 273, 2006
|
|
2AWW
 
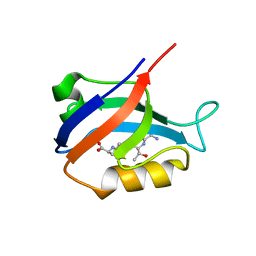 | | Synapse associated protein 97 PDZ2 domain variant C378G with C-terminal GluR-A peptide | | Descriptor: | 18-residue C-terminal peptide from glutamate receptor, ionotropic, AMPA1, ... | | Authors: | Von Ossowski, I, Oksanen, E, Von Ossowski, L, Cai, C, Sundberg, M, Goldman, A, Keinanen, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-08-29 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of the second PDZ domain of SAP97 in complex with a GluR-A C-terminal peptide
Febs J., 273, 2006
|
|
2AWX
 
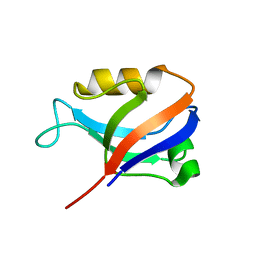 | | Synapse associated protein 97 PDZ2 domain variant C378S | | Descriptor: | HISTIDINE, Synapse associated protein 97 | | Authors: | Von Ossowski, I, Oksanen, E, Von Ossowski, L, Cai, C, Sundberg, M, Goldman, A, Keinanen, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the second PDZ domain of SAP97 in complex with a GluR-A C-terminal peptide
Febs J., 273, 2006
|
|
2AWU
 
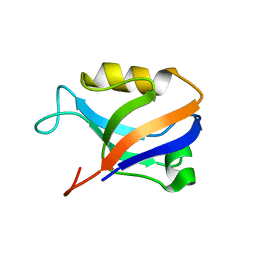 | | Synapse associated protein 97 PDZ2 domain variant C378G | | Descriptor: | AHH, Synapse-associated protein 97 | | Authors: | Von Ossowski, I, Oksanen, E, Von Ossowski, L, Cai, C, Sundberg, M, Goldman, A, Keinanen, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-08-29 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structure of the second PDZ domain of SAP97 in complex with a GluR-A C-terminal peptide
Febs J., 273, 2006
|
|
2V5E
 
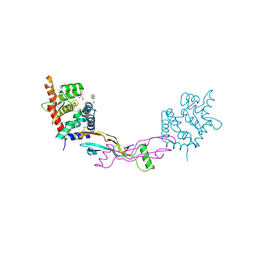 | | The structure of the GDNF:Coreceptor complex: Insights into RET signalling and heparin binding. | | Descriptor: | 1,2-ETHANEDIOL, 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Parkash, V, Leppanen, V.-M, Virtanen, H, Jurvansuu, J.-M, Bespalov, M.M, Sidorova, Y.A, Runeberg-Roos, P, Saarma, M, Goldman, A. | | Deposit date: | 2008-10-03 | | Release date: | 2008-10-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Structure of the Glial Cell Line-Derived Neurotrophic Factor-Coreceptor Complex: Insights Into Ret Signaling and Heparin Binding.
J.Biol.Chem., 283, 2008
|
|
2W50
 
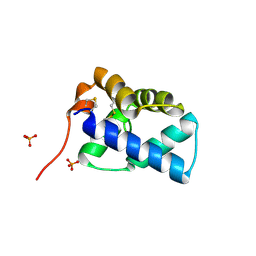 | | N-terminal domain of human conserved dopamine neurotrophic factor (CDNF) | | Descriptor: | ARMET-LIKE PROTEIN 1, PHOSPHATE ION | | Authors: | Parkash, V, Lindholm, P, Peranen, J, Kalkkinen, N, Oksanen, E, Saarma, M, Leppanen, V.M, Goldman, A. | | Deposit date: | 2008-12-03 | | Release date: | 2009-03-17 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure of the Conserved Neurotrophic Factors Manf and Cdnf Explains Why They are Bifunctional.
Protein Eng.Des.Sel., 22, 2009
|
|
2W51
 
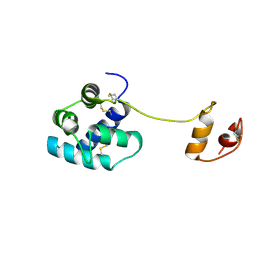 | | Human mesencephalic astrocyte-derived neurotrophic factor (MANF) | | Descriptor: | PROTEIN ARMET | | Authors: | Parkash, V, Lindholm, P, Peranen, J, Kalkkinen, N, Oksanen, E, Saarma, M, Leppanen, V.M, Goldman, A. | | Deposit date: | 2008-12-03 | | Release date: | 2009-03-17 | | Last modified: | 2019-04-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure of the Conserved Neurotrophic Factors Manf and Cdnf Explains Why They are Bifunctional.
Protein Eng.Des.Sel., 22, 2009
|
|
2XOT
 
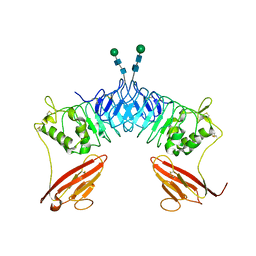 | | Crystal structure of neuronal leucine rich repeat protein AMIGO-1 | | Descriptor: | Amphoterin-induced protein 1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kajander, T, Kuja-Panula, J, Rauvala, H, Goldman, A. | | Deposit date: | 2010-08-23 | | Release date: | 2011-09-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure and Role of Glycans and Dimerisation in Folding of Neuronal Leucine-Rich Repeat Protein Amigo-1
J.Mol.Biol., 413, 2011
|
|
2Y69
 
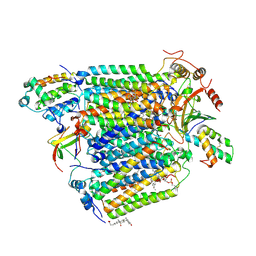 | | Bovine heart cytochrome c oxidase re-refined with molecular oxygen | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, CHOLIC ACID, ... | | Authors: | Kaila, V.R.I, Oksanen, E, Goldman, A, Verkhovsky, M.I, Sundholm, D, Wikstrom, M. | | Deposit date: | 2011-01-20 | | Release date: | 2011-02-23 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Combined Quantum Chemical and Crystallographic Study on the Oxidized Binuclear Center of Cytochrome C Oxidase.
Biochim.Biophys.Acta, 1807, 2011
|
|
1YQ8
 
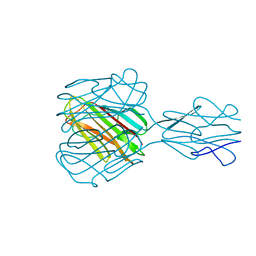 | | PRD1 vertex protein P5 | | Descriptor: | Minor capsid protein | | Authors: | Merckel, M.C, Huiskonen, J.T, Goldman, A, Bamford, D.H, Tuma, R. | | Deposit date: | 2005-02-01 | | Release date: | 2005-04-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of the bacteriophage PRD1 spike sheds light on the evolution of viral capsid architecture.
Mol.Cell, 18, 2005
|
|
