7T6F
 
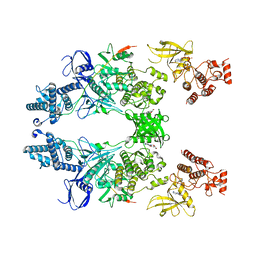 | | Structure of active Janus Kinase (JAK) dimer complexed with cytokine receptor intracellular domain | | Descriptor: | ADENOSINE, ADENOSINE-5'-DIPHOSPHATE, Interferon lambda receptor 1, ... | | Authors: | Glassman, C.R, Tsutsumi, N, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-12-13 | | Release date: | 2022-03-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a Janus kinase cytokine receptor complex reveals the basis for dimeric activation.
Science, 376, 2022
|
|
6WDQ
 
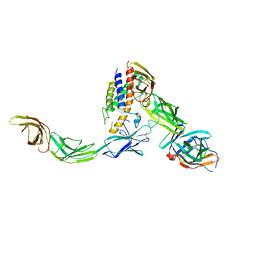 | | IL23/IL23R/IL12Rb1 signaling complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 receptor subunit beta-1, ... | | Authors: | Jude, K.M, Ely, L.K, Glassman, C.R, Thomas, C, Spangler, J.B, Lupardus, P.J, Garcia, K.C. | | Deposit date: | 2020-04-01 | | Release date: | 2021-02-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for IL-12 and IL-23 receptor sharing reveals a gateway for shaping actions on T versus NK cells.
Cell, 184, 2021
|
|
7S2S
 
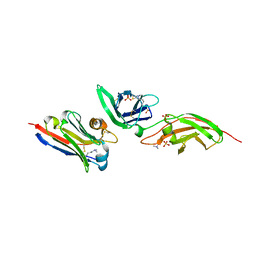 | | nanobody bound to Interleukin-2Rbeta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, IL2Rb-binding nanobody, Interleukin-2 receptor subunit beta, ... | | Authors: | Glassman, C.R, Jude, K.M, Yen, M, Garcia, K.C. | | Deposit date: | 2021-09-03 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Facile discovery of surrogate cytokine agonists.
Cell, 185, 2022
|
|
7S2R
 
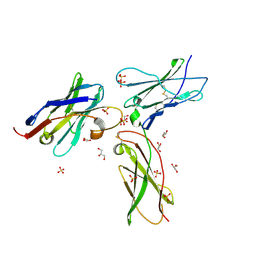 | | nanobody bound to IL-2Rg | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor common subunit gamma, ... | | Authors: | Glassman, C.R, Jude, K.M, Yen, M, Garcia, K.C. | | Deposit date: | 2021-09-03 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Facile discovery of surrogate cytokine agonists.
Cell, 185, 2022
|
|
7U7N
 
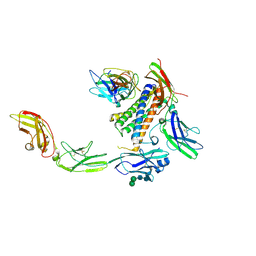 | | IL-27 quaternary receptor signaling complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-27 receptor subunit alpha, ... | | Authors: | Caveney, N.A, Glassman, C.R, Jude, K.M, Tsutsumi, N, Garcia, K.C. | | Deposit date: | 2022-03-07 | | Release date: | 2022-05-25 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structure of the IL-27 quaternary receptor signaling complex.
Elife, 11, 2022
|
|
6WDP
 
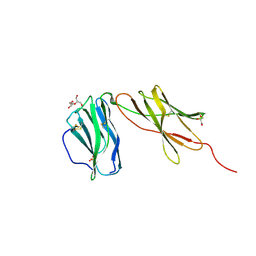 | | Interleukin 12 receptor subunit beta-1 | | Descriptor: | GLYCEROL, Interleukin-12 receptor subunit beta-1, SULFATE ION | | Authors: | Spangler, J.B, Thomas, C, Jude, K.M, Garcia, K.C. | | Deposit date: | 2020-04-01 | | Release date: | 2021-02-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural basis for IL-12 and IL-23 receptor sharing reveals a gateway for shaping actions on T versus NK cells.
Cell, 184, 2021
|
|
8CTG
 
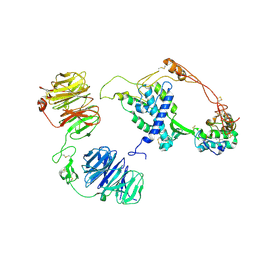 | | Extracellular architecture of an engineered canonical Wnt signaling ternary complex | | Descriptor: | Frizzled-8, Low-density lipoprotein receptor-related protein 6, PALMITOLEIC ACID, ... | | Authors: | Tsutsumi, N, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-05-14 | | Release date: | 2023-03-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Wnt-Frizzled-LRP6 initiation complex reveals the basis for coreceptor discrimination.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6BGA
 
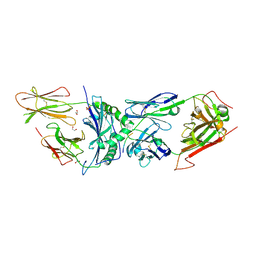 | | 2B4 I-Ek TCR-MHC complex with affinity-enhancing Velcro peptide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2B4 peptide,MHC I-Ek B chain, ... | | Authors: | Gee, M.H, Sibener, L.V, Birnbaum, M.E, Jude, K.M, Garcia, K.C. | | Deposit date: | 2017-10-27 | | Release date: | 2018-07-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | Stress-testing the relationship between T cell receptor/peptide-MHC affinity and cross-reactivity using peptide velcro.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8EWY
 
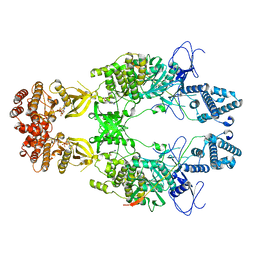 | | Structure of Janus Kinase (JAK) dimer complexed with cytokine receptor intracellular domain | | Descriptor: | ADENOSINE, ADENOSINE-5'-DIPHOSPHATE, Interferon lambda receptor 1, ... | | Authors: | Caveney, N.A, Saxton, R.A, Waghray, D, Garcia, K.C. | | Deposit date: | 2022-10-24 | | Release date: | 2023-03-08 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structural basis of Janus kinase trans-activation.
Cell Rep, 42, 2023
|
|
8FFE
 
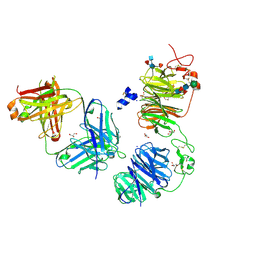 | | Crystal structure of LRP6 E1E2 domains bound to YW210.09 Fab and engineered XWnt8 peptide | | Descriptor: | GLYCEROL, Low-density lipoprotein receptor-related protein 6, SODIUM ION, ... | | Authors: | Jude, K.M, Tsutsumi, N, Waghray, D, Garcia, K.C. | | Deposit date: | 2022-12-08 | | Release date: | 2023-03-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure of the Wnt-Frizzled-LRP6 initiation complex reveals the basis for coreceptor discrimination.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8DA3
 
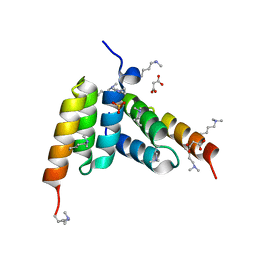 | | Coevolved affibody-Z domain pair LL1.c1 | | Descriptor: | Affibody LL1.FILF, Immunoglobulin G-binding protein A, MALONATE ION, ... | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
8DA9
 
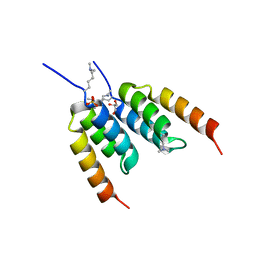 | | Coevolved affibody-Z domain pair LL2.c3 | | Descriptor: | Affibody LL2.FIIV, GLYCEROL, Immunoglobulin G-binding protein A, ... | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
8DA5
 
 | | Coevolved affibody-Z domain pair LL1.c4 | | Descriptor: | GLYCEROL, Immunoglobulin G-binding protein A, affibody LL1.FIVM | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
8DA8
 
 | | Coevolved affibody-Z domain pair LL2.c1 | | Descriptor: | Affibody LL2.FIIK, GLYCEROL, Immunoglobulin G-binding protein A | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
8DAC
 
 | | Coevolved affibody-Z domain pair LL2.c22 | | Descriptor: | Affibody LL2.FILV, GLYCEROL, Immunoglobulin G-binding protein A | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
8DA4
 
 | | Coevolved affibody-Z domain pair LL1.c2 | | Descriptor: | Affibody LL1.FIVM, Immunoglobulin G-binding protein A, SULFATE ION, ... | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
8DA6
 
 | | Coevolved affibody-Z domain pair LL1.c5 | | Descriptor: | Affibody LL1.FIIM, Immunoglobulin G-binding protein A | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
8DA7
 
 | | Coevolved affibody-Z domain pair LL1.c6 | | Descriptor: | Immunoglobulin G-binding protein A, MALONATE ION, affibody LL1.FIFV | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
8DAB
 
 | | Coevolved affibody-Z domain pair LL2.c17 | | Descriptor: | Affibody LL2.IVVY, Immunoglobulin G-binding protein A | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.134 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
8DAA
 
 | | Coevolved affibody-Z domain pair LL2.c7 | | Descriptor: | Affibody LL2.FIVK, Immunoglobulin G-binding protein A, MALONATE ION | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
