1ZY1
 
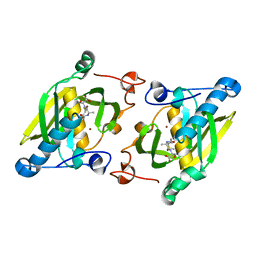 | | X-ray structure of peptide deformylase from Arabidopsis thaliana (AtPDF1A) in complex with Met-Ala-Ser | | Descriptor: | Peptide deformylase, mitochondrial, ZINC ION, ... | | Authors: | Fieulaine, S, Juillan-Binard, C, Serero, A, Dardel, F, Giglione, C, Meinnel, T, Ferrer, J.-L. | | Deposit date: | 2005-06-09 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of mitochondrial (Type 1A) peptide deformylase provides clear guidelines for the design of inhibitors specific for the bacterial forms
J.Biol.Chem., 280, 2005
|
|
6QRM
 
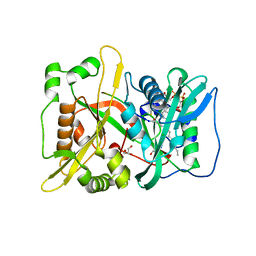 | | HsNMT1 in complex with both MyrCoA and GNCFSKRRAA substrates | | Descriptor: | Apoptosis-inducing factor 3, CHLORIDE ION, COENZYME A, ... | | Authors: | Dian, C, Riviere, F.B, Asensio, T, Giglione, C, Meinnel, T. | | Deposit date: | 2019-02-19 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | High-resolution snapshots of human N-myristoyltransferase in action illuminate a mechanism promoting N-terminal Lys and Gly myristoylation.
Nat Commun, 11, 2020
|
|
1ZXZ
 
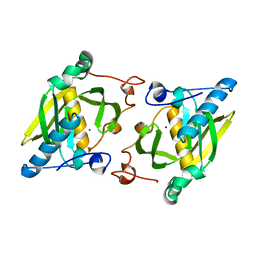 | | X-ray structure of peptide deformylase from Arabidopsis thaliana (AtPDF1A); crystals grown in PEG-5000 MME as precipitant | | Descriptor: | Peptide deformylase, mitochondrial, ZINC ION | | Authors: | Fieulaine, S, Juillan-Binard, C, Serero, A, Dardel, F, Giglione, C, Meinnel, T, Ferrer, J.-L. | | Deposit date: | 2005-06-09 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of mitochondrial (Type 1A) peptide deformylase provides clear guidelines for the design of inhibitors specific for the bacterial forms
J.Biol.Chem., 280, 2005
|
|
1ZY0
 
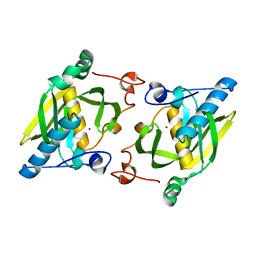 | | X-ray structure of peptide deformylase from Arabidopsis thaliana (AtPDF1A); crystals grown in PEG-6000 | | Descriptor: | Peptide deformylase, mitochondrial, ZINC ION | | Authors: | Fieulaine, S, Juillan-Binard, C, Serero, A, Dardel, F, Giglione, C, Meinnel, T, Ferrer, J.-L. | | Deposit date: | 2005-06-09 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of mitochondrial (Type 1A) peptide deformylase provides clear guidelines for the design of inhibitors specific for the bacterial forms
J.Biol.Chem., 280, 2005
|
|
5JEZ
 
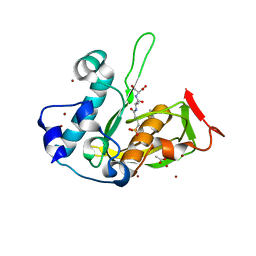 | | Crystal structure of type 2 PDF from Streptococcus agalactiae in complex with tripeptide Met-Ala-Ser | | Descriptor: | ACETATE ION, Met-Ala-Ser, Peptide deformylase, ... | | Authors: | Fieulaine, S, Giglione, C, Meinnel, T. | | Deposit date: | 2016-04-19 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A unique peptide deformylase platform to rationally design and challenge novel active compounds.
Sci Rep, 6, 2016
|
|
5JF7
 
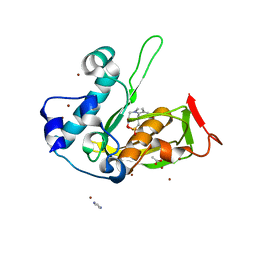 | | Crystal structure of type 2 PDF from Streptococcus agalactiae in complex with inhibitor SMP289 | | Descriptor: | 2-(3-benzyl-5-bromo-1H-indol-1-yl)-N-hydroxyacetamide, ACETATE ION, IMIDAZOLE, ... | | Authors: | Fieulaine, S, Giglione, C, Meinnel, T, Hamiche, K. | | Deposit date: | 2016-04-19 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A unique peptide deformylase platform to rationally design and challenge novel active compounds.
Sci Rep, 6, 2016
|
|
5JF8
 
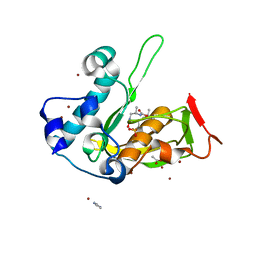 | | Crystal structure of type 2 PDF from Streptococcus agalactiae in complex with inhibitor RAS358 (21) | | Descriptor: | ACETATE ION, IMIDAZOLE, Peptide deformylase, ... | | Authors: | Fieulaine, S, Giglione, C, Meinnel, T. | | Deposit date: | 2016-04-19 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A unique peptide deformylase platform to rationally design and challenge novel active compounds.
Sci Rep, 6, 2016
|
|
1LRY
 
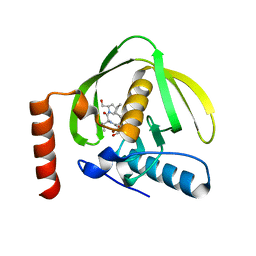 | | Crystal Structure of P. aeruginosa Peptide Deformylase Complexed with Antibiotic Actinonin | | Descriptor: | ACTINONIN, PEPTIDE deformylase, ZINC ION | | Authors: | Guilloteau, J.-P, Mathieu, M, Giglione, C, Blanc, V, Dupuy, A, Chevrier, M, Gil, P, Famechon, A, Meinnel, T, Mikol, V. | | Deposit date: | 2002-05-16 | | Release date: | 2002-07-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structures of four peptide deformylases bound to the antibiotic actinonin reveal two distinct types: a platform for the structure-based design of antibacterial agents.
J.Mol.Biol., 320, 2002
|
|
4JE6
 
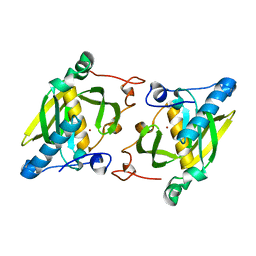 | | Crystal structure of a human-like mitochondrial peptide deformylase | | Descriptor: | Peptide deformylase 1A, chloroplastic/mitochondrial, ZINC ION | | Authors: | Fieulaine, S, Meinnel, T, Giglione, C. | | Deposit date: | 2013-02-26 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Understanding the highly efficient catalysis of prokaryotic peptide deformylases by shedding light on the determinants specifying the low activity of the human counterpart.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1LRU
 
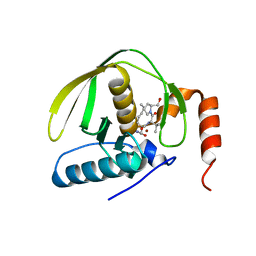 | | Crystal Structure of E.coli Peptide Deformylase Complexed with Antibiotic Actinonin | | Descriptor: | ACTINONIN, PEPTIDE DEFORMYLASE, SULFATE ION, ... | | Authors: | Guilloteau, J.-P, Mathieu, M, Giglione, C, Blanc, V, Dupuy, A, Chevrier, M, Gil, P, Famechon, A, Meinnel, T, Mikol, V. | | Deposit date: | 2002-05-16 | | Release date: | 2002-07-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structures of four peptide deformylases bound to the antibiotic actinonin reveal two distinct types: a platform for the structure-based design of antibacterial agents.
J.Mol.Biol., 320, 2002
|
|
4JE8
 
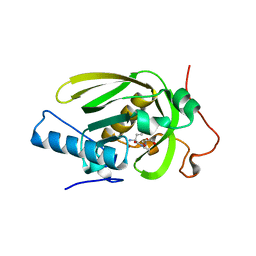 | | Crystal structure of a human-like mitochondrial peptide deformylase in complex with Met-Ala-Ser | | Descriptor: | Peptide deformylase 1A, chloroplastic/mitochondrial, ZINC ION, ... | | Authors: | Fieulaine, S, Meinnel, T, Giglione, C. | | Deposit date: | 2013-02-26 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Understanding the highly efficient catalysis of prokaryotic peptide deformylases by shedding light on the determinants specifying the low activity of the human counterpart.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4JE7
 
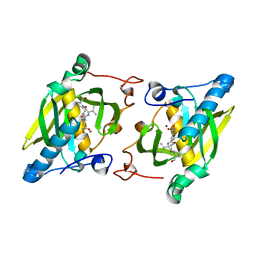 | | Crystal structure of a human-like mitochondrial peptide deformylase in complex with actinonin | | Descriptor: | ACTINONIN, Peptide deformylase 1A, chloroplastic/mitochondrial, ... | | Authors: | Fieulaine, S, Meinnel, T, Giglione, C. | | Deposit date: | 2013-02-26 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Understanding the highly efficient catalysis of prokaryotic peptide deformylases by shedding light on the determinants specifying the low activity of the human counterpart.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8Q26
 
 | | HsNMT1 in complex with both MyrCoA and GNCFSKPR inhibitor peptide | | Descriptor: | CHLORIDE ION, COENZYME A, Glycylpeptide N-tetradecanoyltransferase 1, ... | | Authors: | Dian, C, Giglione, C, Meinnel, T. | | Deposit date: | 2023-08-01 | | Release date: | 2024-08-14 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel, tightly structurally related N-myristoyltransferase inhibitors display equally potent yet distinct inhibitory mechanisms.
Structure, 32, 2024
|
|
8Q3D
 
 | | HsNMT1 in complex with both MyrCoA and GNCFSKPR(NH2) inhibitor peptide | | Descriptor: | CHLORIDE ION, COENZYME A, GLYCEROL, ... | | Authors: | Dian, C, Giglione, C, Meinnel, T. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-14 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Novel, tightly structurally related N-myristoyltransferase inhibitors display equally potent yet distinct inhibitory mechanisms.
Structure, 32, 2024
|
|
8Q3S
 
 | | HsNMT1 in complex with both MyrCoA and GNCFSKAR inhibitor peptide | | Descriptor: | CHLORIDE ION, COENZYME A, GLY-ASN-CYS-PHE-SER-LYS-ALA-ARG, ... | | Authors: | Dian, C, Giglione, C, Meinnel, T. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-14 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Novel, tightly structurally related N-myristoyltransferase inhibitors display equally potent yet distinct inhibitory mechanisms.
Structure, 32, 2024
|
|
8Q23
 
 | | HsNMT1 in complex with both MyrCoA and Ac-D-ORN-SFSKPR inhibitor peptide | | Descriptor: | ACE-ORD-SER-PHE-SER-LYS-PRO-ARG, CHLORIDE ION, GLYCEROL, ... | | Authors: | Dian, C, Giglione, C, Meinnel, T. | | Deposit date: | 2023-08-01 | | Release date: | 2024-08-14 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel, tightly structurally related N-myristoyltransferase inhibitors display equally potent yet distinct inhibitory mechanisms.
Structure, 32, 2024
|
|
8Q3T
 
 | | HsNMT1 in complex with both MyrCoA and GNCFSKPRVPTK inhibitor peptide | | Descriptor: | CHLORIDE ION, COENZYME A, GLYCEROL, ... | | Authors: | Dian, C, Giglione, C, Meinnel, T. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-14 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Novel, tightly structurally related N-myristoyltransferase inhibitors display equally potent yet distinct inhibitory mechanisms.
Structure, 32, 2024
|
|
8Q2Z
 
 | | HsNMT1 in complex with both MyrCoA and GNLLSKFR peptide | | Descriptor: | CHLORIDE ION, COENZYME A, GLYCEROL, ... | | Authors: | Dian, C, Giglione, C, Meinnel, T. | | Deposit date: | 2023-08-03 | | Release date: | 2024-08-14 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Novel, tightly structurally related N-myristoyltransferase inhibitors display equally potent yet distinct inhibitory mechanisms.
Structure, 32, 2024
|
|
8Q24
 
 | | HsNMT1 in complex with both MyrCoA and (DAB)SFSKPR inhibitor peptide | | Descriptor: | CHLORIDE ION, DAB-SER-PHE-SER-LYS-PRO-ARG, GLYCEROL, ... | | Authors: | Dian, C, Giglione, C, Meinnel, T. | | Deposit date: | 2023-08-01 | | Release date: | 2024-08-14 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel, tightly structurally related N-myristoyltransferase inhibitors display equally potent yet distinct inhibitory mechanisms.
Structure, 32, 2024
|
|
7OWQ
 
 | |
7OWU
 
 | | HsNMT1 in complex with both CoA and Myr-ANCFSKPR peptide | | Descriptor: | ALA-ASN-CYS-PHE-SER-LYS-PRO-ARG, COENZYME A, GLYCEROL, ... | | Authors: | Dian, C, Giglione, C, Meinnel, T. | | Deposit date: | 2021-06-18 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural and Large-scale Analysis Unveil the Intertwined Paths Promoting NMT-catalyzed Lysine and Glycine Myristoylation.
J.Mol.Biol., 434, 2022
|
|
7OWO
 
 | | HsNMT1 in complex with both MyrCoA and N-acetylated KSFSKPR peptide | | Descriptor: | COENZYME A, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase 1, ... | | Authors: | Dian, C, Giglione, C, Meinnel, T. | | Deposit date: | 2021-06-18 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Large-scale Analysis Unveil the Intertwined Paths Promoting NMT-catalyzed Lysine and Glycine Myristoylation.
J.Mol.Biol., 434, 2022
|
|
7OWR
 
 | | HsNMT1 in complex with both MyrCoA and peptide GGKSFSKPR | | Descriptor: | GLY-GLY-LYS-SER-PHE-SER-LYS-PRO-ARG, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase 1, ... | | Authors: | Dian, C, Giglione, C, Meinnel, T. | | Deposit date: | 2021-06-18 | | Release date: | 2022-12-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural and Large-scale Analysis Unveil the Intertwined Paths Promoting NMT-catalyzed Lysine and Glycine Myristoylation.
J.Mol.Biol., 434, 2022
|
|
7OWN
 
 | | HsNMT1 in complex with both MyrCoA and peptide AKSFSKPR | | Descriptor: | ALA-LYS-SER-PHE-SER-LYS-PRO-ARG, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase 1, ... | | Authors: | Dian, C, Giglione, C, Meinnel, T. | | Deposit date: | 2021-06-18 | | Release date: | 2022-12-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Large-scale Analysis Unveil the Intertwined Paths Promoting NMT-catalyzed Lysine and Glycine Myristoylation.
J.Mol.Biol., 434, 2022
|
|
7OWM
 
 | | HsNMT1 in complex with both MyrCoA and HCPA substrate peptide GKQNSKLR | | Descriptor: | GLYCEROL, Glycylpeptide N-tetradecanoyltransferase 1, Neuron-specific calcium-binding protein hippocalcin, ... | | Authors: | Dian, C, Giglione, C, Meinnel, T. | | Deposit date: | 2021-06-18 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Large-scale Analysis Unveil the Intertwined Paths Promoting NMT-catalyzed Lysine and Glycine Myristoylation.
J.Mol.Biol., 434, 2022
|
|
