3O10
 
 | | Crystal structure of the HEPN domain from human sacsin | | Descriptor: | MALONATE ION, Sacsin | | Authors: | Kozlov, G, Gehring, K. | | Deposit date: | 2010-07-20 | | Release date: | 2011-03-30 | | Last modified: | 2014-07-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of defects in the sacsin HEPN domain responsible for autosomal recessive spastic ataxia of Charlevoix-Saguenay (ARSACS).
J.Biol.Chem., 286, 2011
|
|
2OOA
 
 | |
2OO9
 
 | |
2OOB
 
 | |
5TDB
 
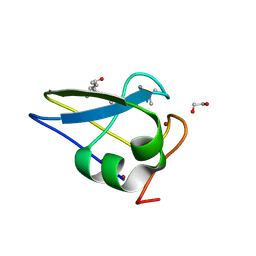 | | Crystal structure of the human UBR-box domain from UBR2 in complex with asymmetrically double methylated arginine peptide | | Descriptor: | 1,2-ETHANEDIOL, DA2-ILE-PHE-SER peptide, E3 ubiquitin-protein ligase UBR2, ... | | Authors: | Munoz-Escobar, J, Kozlov, G, Gehring, K. | | Deposit date: | 2016-09-19 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Bound Waters Mediate Binding of Diverse Substrates to a Ubiquitin Ligase.
Structure, 25, 2017
|
|
5TDC
 
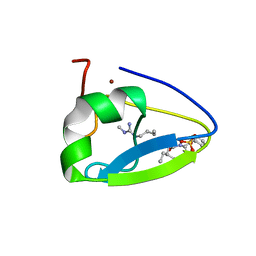 | | Crystal structure of the human UBR-box domain from UBR1 in complex with monomethylated arginine peptide. | | Descriptor: | E3 ubiquitin-protein ligase UBR1, NMM-ILE-PHE-SER peptide, SULFATE ION, ... | | Authors: | Kozlov, G, Munoz-Escobar, J, Matta-Camacho, E, Gehring, K. | | Deposit date: | 2016-09-19 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.607 Å) | | Cite: | Bound Waters Mediate Binding of Diverse Substrates to a Ubiquitin Ligase.
Structure, 25, 2017
|
|
5TDD
 
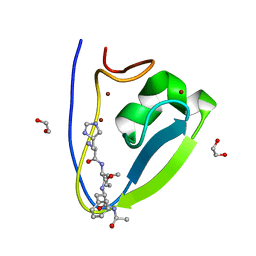 | | Human UBR-box from UBR2 in complex with HIFS peptide | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase UBR2, HIS-ILE-PHE-SER peptide, ... | | Authors: | Munoz-Escobar, J, Kozlov, G, Gehring, K. | | Deposit date: | 2016-09-19 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Bound Waters Mediate Binding of Diverse Substrates to a Ubiquitin Ligase.
Structure, 25, 2017
|
|
5TGG
 
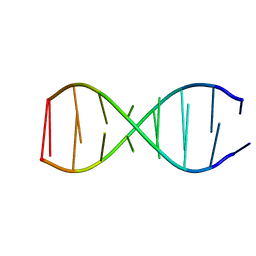 | |
5TSR
 
 | |
5UM3
 
 | |
5V46
 
 | | Crystal structure of the I113M, F270M, K291M, L308M mutant of SR1 domain of human sacsin | | Descriptor: | Sacsin | | Authors: | Menade, M, Kozlov, G, Gehring, K. | | Deposit date: | 2017-03-08 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of ubiquitin-like (Ubl) and Hsp90-like domains of sacsin provide insight into pathological mutations.
J. Biol. Chem., 293, 2018
|
|
5V45
 
 | |
5V47
 
 | | Crystal structure of the SR1 domain of lizard sacsin | | Descriptor: | Lizard sacsin, SULFATE ION | | Authors: | Pan, T, Menade, M, Kozlov, G, Gehring, K. | | Deposit date: | 2017-03-08 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structures of ubiquitin-like (Ubl) and Hsp90-like domains of sacsin provide insight into pathological mutations.
J. Biol. Chem., 293, 2018
|
|
5V44
 
 | |
5VXQ
 
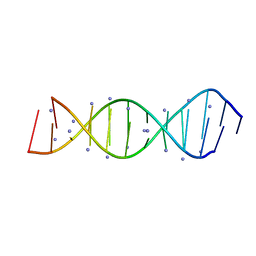 | | X-Ray crystallography structure of the parallel stranded duplex formed by 5-rA5-dA-rA5 | | Descriptor: | AMMONIUM ION, DNA/RNA (5'-R(*AP*AP*AP*AP*A)-D(P*A)-R(P*AP*AP*AP*AP*A)-3') | | Authors: | Xie, J, Chen, Y, Wei, X, Kozlov, G, Gehring, K. | | Deposit date: | 2017-05-23 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.002 Å) | | Cite: | Influence of nucleotide modifications at the C2' position on the Hoogsteen base-paired parallel-stranded duplex of poly(A) RNA.
Nucleic Acids Res., 45, 2017
|
|
5VMD
 
 | | Crystal structure of UBR-box from UBR6 in a domain-swapping conformation | | Descriptor: | 1,2-ETHANEDIOL, F-box only protein 11, ZINC ION | | Authors: | Munoz-Escobar, J, Kozlov, G, Gehring, K. | | Deposit date: | 2017-04-27 | | Release date: | 2017-07-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Crystal structure of the UBR-box from UBR6/FBXO11 reveals domain swapping mediated by zinc binding.
Protein Sci., 26, 2017
|
|
5VSZ
 
 | |
5VSX
 
 | |
1RRZ
 
 | |
1R6H
 
 | | Solution Structure of human PRL-3 | | Descriptor: | protein tyrosine phosphatase type IVA, member 3 isoform 1 | | Authors: | Kozlov, G, Gehring, K, Ekiel, I. | | Deposit date: | 2003-10-15 | | Release date: | 2004-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Insights into Molecular Function of the Metastasis-associated Phosphatase PRL-3.
J.Biol.Chem., 279, 2004
|
|
1RWU
 
 | |
1RYJ
 
 | | Solution NMR Structure of Protein Mth1743 from Methanobacterium thermoautotrophicum. Ontario Centre for Structural Proteomics target MTH1743_1_70; Northeast Structural Genomics Consortium Target TT526. | | Descriptor: | unknown | | Authors: | Yee, A, Chang, X, Pineda-Lucena, A, Wu, B, Semesi, A, Le, B, Ramelot, T, Lee, G.M, Bhattacharyya, S, Gutierrez, P, Denisov, A, Lee, C.H, Cort, J.R, Kozlov, G, Liao, J, Finak, G, Chen, L, Wishart, D, Lee, W, McIntosh, L.P, Gehring, K, Kennedy, M.A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-12-22 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | AN NMR APPROACH TO STRUCTURAL PROTEOMICS
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1SG5
 
 | |
1SG7
 
 | | NMR solution structure of the putative cation transport regulator ChaB | | Descriptor: | Putative Cation transport regulator chaB | | Authors: | Osborne, M.J, Siddiqui, N, Cygler, M, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2004-02-23 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of ChaB, a putative membrane ion antiporter regulator from Escherichia coli
BMC STRUCT.BIOL., 4, 2004
|
|
1SSL
 
 | | Solution structure of the PSI domain from the Met receptor | | Descriptor: | Hepatocyte growth factor receptor | | Authors: | Kozlov, G, Perreault, A, Schrag, J.D, Cygler, M, Gehring, K, Ekiel, I. | | Deposit date: | 2004-03-24 | | Release date: | 2004-10-12 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Insights into function of PSI domains from structure of the Met receptor PSI domain.
Biochem.Biophys.Res.Commun., 321, 2004
|
|
