1PPN
 
 | | STRUCTURE OF MONOCLINIC PAPAIN AT 1.60 ANGSTROMS RESOLUTION | | Descriptor: | METHANOL, PAPAIN, UNKNOWN LIGAND | | Authors: | Pickersgill, R.W, Harris, G.W, Garman, E. | | Deposit date: | 1991-10-25 | | Release date: | 1994-01-31 | | Last modified: | 2015-04-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Monoclinic Papain at 1.60 Angstroms Resolution
Acta Crystallogr.,Sect.B, 48, 1992
|
|
1Q27
 
 | |
1B64
 
 | | SOLUTION STRUCTURE OF THE GUANINE NUCLEOTIDE EXCHANGE FACTOR DOMAIN FROM HUMAN ELONGATION FACTOR-ONE BETA, NMR, 20 STRUCTURES | | Descriptor: | ELONGATION FACTOR 1-BETA | | Authors: | Perez, J.M.J, Siegal, G, Kriek, J, Hard, K, Dijk, J, Canters, G.W, Moller, W. | | Deposit date: | 1999-01-20 | | Release date: | 1999-05-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the guanine nucleotide exchange domain of human elongation factor 1beta reveals a striking resemblance to that of EF-Ts from Escherichia coli.
Structure Fold.Des., 7, 1999
|
|
1C8P
 
 | | NMR STRUCTURE OF THE LIGAND BINDING DOMAIN OF THE COMMON BETA-CHAIN IN THE GM-CSF, IL-3 AND IL-5 RECEPTORS | | Descriptor: | CYTOKINE RECEPTOR COMMON BETA CHAIN | | Authors: | Mulhern, T.D, D'Andrea, R.J, Gaunt, C, Vandeleur, L, Vadas, M.A, Lopez, A.F, Booker, G.W, Bagley, C.J. | | Deposit date: | 1999-10-05 | | Release date: | 2000-06-15 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the cytokine-binding domain of the common beta-chain of the receptors for granulocyte-macrophage colony-stimulating factor, interleukin-3 and interleukin-5.
J.Mol.Biol., 297, 2000
|
|
1CLX
 
 | | CATALYTIC CORE OF XYLANASE A | | Descriptor: | CALCIUM ION, XYLANASE A | | Authors: | Harris, G.W, Jenkins, J.A, Connerton, I, Pickersgill, R.W. | | Deposit date: | 1995-08-31 | | Release date: | 1996-06-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined crystal structure of the catalytic domain of xylanase A from Pseudomonas fluorescens at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
7TVL
 
 | | Viral AMG chitosanase V-Csn, apo structure | | Descriptor: | GLYCEROL, SULFATE ION, Viral chitosanase V-Csn | | Authors: | Smith, C.A, Wu, R, Buchko, G.W, Cort, J.R, Hofmockel, K.S, Jansson, J.K. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Structural characterization of a soil viral auxiliary metabolic gene product - a functional chitosanase.
Nat Commun, 13, 2022
|
|
7TVN
 
 | | Viral AMG chitosanase V-Csn, D148N mutant | | Descriptor: | GLYCEROL, SULFATE ION, Viral chitosanase V-Csn D148N mutant | | Authors: | Smith, C.A, Wu, R, Buchko, G.W, Cort, J.R, Hofmockel, K.S, Jansson, J.K. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural characterization of a soil viral auxiliary metabolic gene product - a functional chitosanase.
Nat Commun, 13, 2022
|
|
7TVP
 
 | | Viral AMG chitosanase V-Csn, E157Q mutant, chitotriose complex | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, GLYCEROL, Viral chitosanase V-Csn E157Q mutant chitotriose complex | | Authors: | Smith, C.A, Wu, R, Buchko, G.W, Cort, J.R, Hofmockel, K.S, Jansson, J.K. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural characterization of a soil viral auxiliary metabolic gene product - a functional chitosanase.
Nat Commun, 13, 2022
|
|
7TVO
 
 | | Viral AMG chitosanase V-Csn, E157Q mutant | | Descriptor: | GLYCEROL, SULFATE ION, Viral chitosanase V-Csn E157Q mutant | | Authors: | Smith, C.A, Wu, R, Buchko, G.W, Cort, J.R, Hofmockel, K.S, Jansson, J.K. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural characterization of a soil viral auxiliary metabolic gene product - a functional chitosanase.
Nat Commun, 13, 2022
|
|
7TVM
 
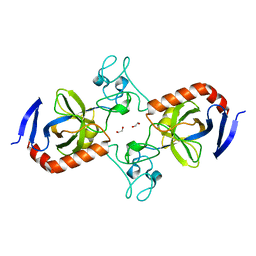 | | Viral AMG chitosanase V-Csn, apo structure, crystal form 2 | | Descriptor: | 1,2-ETHANEDIOL, Viral chitosanase V-Csn | | Authors: | Smith, C.A, Wu, R, Buchko, G.W, Cort, J.R, Hofmockel, K.S, Jansson, J.K. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural characterization of a soil viral auxiliary metabolic gene product - a functional chitosanase.
Nat Commun, 13, 2022
|
|
7TOD
 
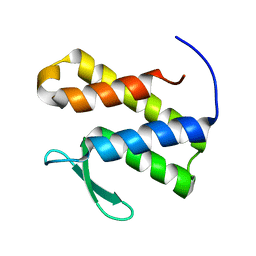 | | Solution structure of the phosphatidylinositol 3-phosphate binding domain from the Legionella effector SetA | | Descriptor: | Subversion of eukaryotic traffic protein A | | Authors: | Beck, W.H.J, Enoki, T.A, Feigenson, G.W, Nicholson, L, Oswald, R, Mao, Y. | | Deposit date: | 2022-01-24 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the phosphatidylinositol 3-phosphate binding domain from the Legionella effector SetA
To Be Published
|
|
7TXX
 
 | |
7XK2
 
 | | Cryo-EM Structure of Human Niacin Receptor HCA2-Gi protein complex | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, Y, Kang, H.J, Gao, R.G, Wang, J.J, Han, G.W, DiBerto, J.F, Wu, L.J, Tong, J.H, Qu, L, Wu, Y.R, Pileski, R, Li, X.M, Zhang, X.C, Zhao, S.W, Kenakin, T, Wang, Q, Stevens, R.C, Peng, W, Roth, B.L, Rao, Z.H, Liu, Z.J. | | Deposit date: | 2022-04-19 | | Release date: | 2023-02-22 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the human niacin receptor HCA2-G i signalling complex.
Nat Commun, 14, 2023
|
|
6W25
 
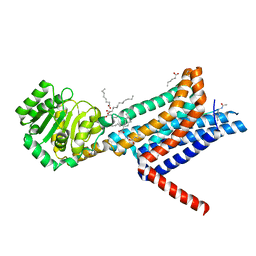 | | Crystal structure of the Melanocortin-4 Receptor (MC4R) in complex with SHU9119 | | Descriptor: | CALCIUM ION, Melanocortin receptor 4,GlgA glycogen synthase,Melanocortin receptor 4, OLEIC ACID, ... | | Authors: | Yu, J, Gimenez, L.E, Hernandez, C.C, Wu, Y, Wein, A.H, Han, G.W, McClary, K, Mittal, S.R, Burdsall, K, Stauch, B, Wu, L, Stevens, S.N, Peisley, A, Williams, S.Y, Chen, V, Millhauser, G.L, Zhao, S, Cone, R.D, Stevens, R.C. | | Deposit date: | 2020-03-04 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Determination of the melanocortin-4 receptor structure identifies Ca2+as a cofactor for ligand binding.
Science, 368, 2020
|
|
6VJM
 
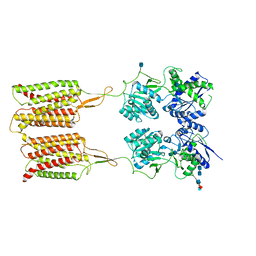 | | Human metabotropic GABA(B) receptor in its apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shaye, H, Han, G.W, Gati, C, Cherezov, V. | | Deposit date: | 2020-01-16 | | Release date: | 2020-06-10 | | Last modified: | 2020-08-26 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Structural basis of the activation of a metabotropic GABA receptor.
Nature, 584, 2020
|
|
6WQA
 
 | | 2.0A angstrom A2a adenosine receptor structure using XFEL data collected in helium atmosphere. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Lee, M.-Y, Geiger, J, Ishchenko, A, Han, G.W, Barty, A, White, T.A, Gati, C, Batyuk, A, Hunter, M.S, Aquila, A, Boutet, S, Weierstall, U, Cherezov, V, Liu, W. | | Deposit date: | 2020-04-28 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Harnessing the power of an X-ray laser for serial crystallography of membrane proteins crystallized in lipidic cubic phase
Iucrj, 7, 2020
|
|
6X94
 
 | | An orthogonal seryl-tRNA synthetase/tRNA pair for noncanonical amino acid mutagenesis in Escherichia coli | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zambaldo, C, Koh, M, Nasertorabi, F, Han, G.W, Chatterjee, A, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2020-06-02 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | An orthogonal seryl-tRNA synthetase/tRNA pair for noncanonical amino acid mutagenesis in Escherichia coli.
Bioorg.Med.Chem., 28, 2020
|
|
2IDF
 
 | | P. aeruginosa azurin N42C/M64E double mutant, BMME-linked dimer | | Descriptor: | 1-[PYRROL-1-YL-2,5-DIONE-METHOXYMETHYL]-PYRROLE-2,5-DIONE, Azurin, COPPER (II) ION, ... | | Authors: | Einsle, O, de Jongh, T.E, Hoffmann, M, Cavazzini, D, Rossi, G.L, Ubbink, M, Canters, G.W. | | Deposit date: | 2006-09-15 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Electron transfer in a crosslinked protein dimer mediated by a hydrogen-bonded network across the dimer interface
To be Published
|
|
7N8O
 
 | | High-resolution structure of photosystem II from the mesophilic cyanobacterium, Synechocystis sp. PCC 6803 | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Gisriel, C.J, Brudvig, G.W. | | Deposit date: | 2021-06-15 | | Release date: | 2021-12-29 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (1.93 Å) | | Cite: | High-resolution cryo-electron microscopy structure of photosystem II from the mesophilic cyanobacterium, Synechocystis sp. PCC 6803.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7OKF
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 8c | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[[4-(4-methylpiperazin-1-yl)-2-oxidanylidene-1H-quinolin-6-yl]amino]pyridine-3-carbonitrile, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Into Deep Water: Optimizing BCL6 Inhibitors by Growing into a Solvated Pocket.
J.Med.Chem., 64, 2021
|
|
7OKI
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 12b | | Descriptor: | (2R)-2-[[6-[(2-chloranyl-3-cyano-pyridin-4-yl)amino]-2-oxidanylidene-1H-quinolin-4-yl]amino]-N-methyl-propanamide, 1,2-ETHANEDIOL, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Into Deep Water: Optimizing BCL6 Inhibitors by Growing into a Solvated Pocket.
J.Med.Chem., 64, 2021
|
|
7OKG
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 8e | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[[4-(1-methylpyrazol-4-yl)-2-oxidanylidene-1H-quinolin-6-yl]amino]pyridine-3-carbonitrile, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Into Deep Water: Optimizing BCL6 Inhibitors by Growing into a Solvated Pocket.
J.Med.Chem., 64, 2021
|
|
7OKK
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 12e | | Descriptor: | 1,2-ETHANEDIOL, 2-[[6-[(2-chloranyl-3-cyano-pyridin-4-yl)amino]-2-oxidanylidene-1H-quinolin-4-yl]amino]-N-methyl-ethanamide, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Into Deep Water: Optimizing BCL6 Inhibitors by Growing into a Solvated Pocket.
J.Med.Chem., 64, 2021
|
|
7OKE
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[(1-methyl-2-oxidanylidene-quinolin-6-yl)amino]pyridine-3-carbonitrile, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Into Deep Water: Optimizing BCL6 Inhibitors by Growing into a Solvated Pocket.
J.Med.Chem., 64, 2021
|
|
7OKH
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 8f | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[[4-(ethylamino)-2-oxidanylidene-1H-quinolin-6-yl]amino]pyridine-3-carbonitrile, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Into Deep Water: Optimizing BCL6 Inhibitors by Growing into a Solvated Pocket.
J.Med.Chem., 64, 2021
|
|
