4QUZ
 
 | |
4QVA
 
 | |
4QVJ
 
 | |
4QV2
 
 | |
2RGV
 
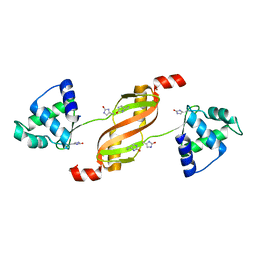 | |
1ZGA
 
 | | Crystal structure of isoflavanone 4'-O-methyltransferase complexed with (+)-6a-hydroxymaackiain | | Descriptor: | (6AR,12AR)-6H-[1,3]DIOXOLO[5,6][1]BENZOFURO[3,2-C]CHROMENE-3,6A(12AH)-DIOL, Isoflavanone 4'-O-methyltransferase', S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Liu, C.-J, Deavours, B.E, Richard, S, Ferrer, J.-L, Dixon, R.A, Noel, J.P. | | Deposit date: | 2005-04-20 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for dual functionality of isoflavonoid O-methyltransferases in the evolution of plant defense responses.
Plant Cell, 18, 2006
|
|
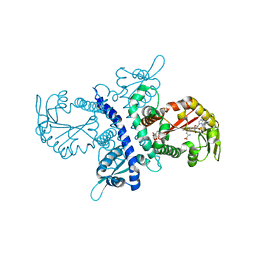
1ZHF
 
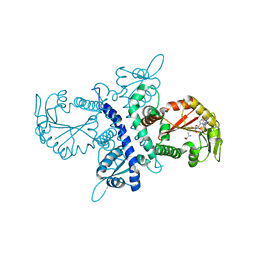 | | Crystal structure of selenomethionine substituted isoflavanone 4'-O-methyltransferase | | Descriptor: | Isoflavanone 4'-O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Liu, C.-J, Deavours, B.E, Richard, S, Ferrer, J.-L, Dixon, R.A, Noel, J.P. | | Deposit date: | 2005-04-25 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for dual functionality of isoflavonoid O-methyltransferases in the evolution of plant defense responses.
Plant Cell, 18, 2006
|
|
1ZG3
 
 | | Crystal structure of the isoflavanone 4'-O-methyltransferase complexed with SAH and 2,7,4'-trihydroxyisoflavanone | | Descriptor: | (2S,3R)-2,7-DIHYDROXY-3-(4-HYDROXYPHENYL)-2,3-DIHYDRO-4H-CHROMEN-4-ONE, S-ADENOSYL-L-HOMOCYSTEINE, isoflavanone 4'-O-methyltransferase | | Authors: | Liu, C.-J, Deavours, B.E, Richard, S, Ferrer, J.-L, Dixon, R.A, Noel, J.P. | | Deposit date: | 2005-04-20 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for dual functionality of isoflavonoid O-methyltransferases in the evolution of plant defense responses.
Plant Cell, 18, 2006
|
|
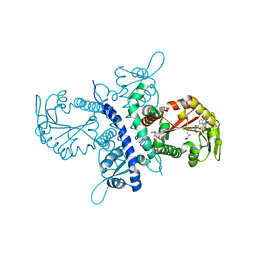
1ZGJ
 
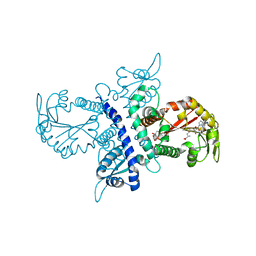 | | Crystal structure of isoflavanone 4'-O-methyltransferase complexed with (+)-pisatin | | Descriptor: | (6AR,12AR)-3-(HYDROXYMETHYL)-6H-[1,3]DIOXOLO[5,6][1]BENZOFURO[3,2-C]CHROMEN-6A(12AH)-OL, Isoflavanone 4'-O-methyltransferase', S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Liu, C.-J, Deavours, B.E, Richard, S, Ferrer, J.-L, Dixon, R.A, Noel, J.P. | | Deposit date: | 2005-04-21 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for dual functionality of isoflavonoid O-methyltransferases in the evolution of plant defense responses.
Plant Cell, 18, 2006
|
|
4ZSD
 
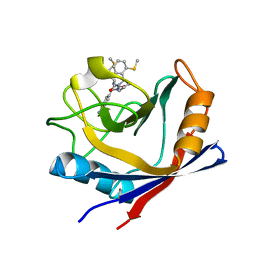 | | Human Cyclophilin D Complexed with an Inhibitor at room temperature | | Descriptor: | 1-(4-aminobenzyl)-3-[(2S)-4-(methylsulfanyl)-1-{(2R)-2-[2-(methylsulfanyl)phenyl]pyrrolidin-1-yl}-1-oxobutan-2-yl]urea, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Gelin, M, Delfosse, V, Allemand, F, Hoh, F, Sallaz-Damaz, Y, Pirocchi, M, Bourguet, W, Ferrer, J.-L, Labesse, G, Guichou, J.-F. | | Deposit date: | 2015-05-13 | | Release date: | 2015-08-12 | | Last modified: | 2015-08-19 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Combining `dry' co-crystallization and in situ diffraction to facilitate ligand screening by X-ray crystallography.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4ZSC
 
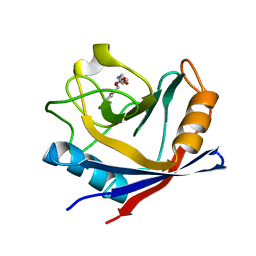 | | Human Cyclophilin D Complexed with an Inhibitor at room temperature | | Descriptor: | Peptidyl-prolyl cis-trans isomerase F, mitochondrial, ethyl N-[(4-aminobenzyl)carbamoyl]glycinate | | Authors: | Gelin, M, Delfosse, V, Allemand, F, Hoh, F, Sallaz-Damaz, Y, Pirocchi, M, Bourguet, W, Ferrer, J.-L, Labesse, G, Guichou, J.-F. | | Deposit date: | 2015-05-13 | | Release date: | 2015-08-12 | | Last modified: | 2015-08-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Combining `dry' co-crystallization and in situ diffraction to facilitate ligand screening by X-ray crystallography.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3CX3
 
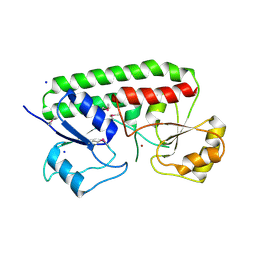 | | Crystal structure Analysis of the Streptococcus pneumoniae AdcAII protein | | Descriptor: | Lipoprotein, SODIUM ION, ZINC ION | | Authors: | Loisel, E, Durmort, C, Jacquamet, L. | | Deposit date: | 2008-04-23 | | Release date: | 2008-08-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | AdcAII, a new pneumococcal Zn-binding protein homologous with ABC transporters: biochemical and structural analysis.
J.Mol.Biol., 381, 2008
|
|
1ODF
 
 | | Structure of YGR205w protein. | | Descriptor: | GLYCEROL, HYPOTHETICAL 33.3 KDA PROTEIN IN ADE3-SER2 INTERGENIC REGION, SULFATE ION | | Authors: | Li De La Sierra-Gallay, I, Van Tilbeurgh, H. | | Deposit date: | 2003-02-19 | | Release date: | 2003-12-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the Ygr205W Protein from Saccharomyces Cerevisiae: Close Structural Resemblance to E.Coli Pantothenate Kinase
Proteins: Struct.,Funct., Genet., 54, 2004
|
|
1QVZ
 
 | | Crystal structure of the S. cerevisiae YDR533c protein | | Descriptor: | YDR533c protein | | Authors: | Graille, M, Leulliot, N, Quevillon-Cheruel, S, van Tilbeurgh, H. | | Deposit date: | 2003-08-29 | | Release date: | 2004-03-30 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the YDR533c S. cerevisiae protein, a class II member of the Hsp31 family
STRUCTURE, 12, 2004
|
|
1QVV
 
 | | Crystal structure of the S. cerevisiae YDR533c protein | | Descriptor: | YDR533c protein | | Authors: | Graille, M, Leulliot, N, Quevillon-Cheruel, S, van Tilbeurgh, H. | | Deposit date: | 2003-08-29 | | Release date: | 2004-03-30 | | Last modified: | 2020-07-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the YDR533c S. cerevisiae protein, a class II member of the Hsp31 family
STRUCTURE, 12, 2004
|
|
1QVW
 
 | | Crystal structure of the S. cerevisiae YDR533c protein | | Descriptor: | GLYCEROL, YDR533c protein | | Authors: | Graille, M, Leulliot, N, Quevillon-Cheruel, S, van Tilbeurgh, H. | | Deposit date: | 2003-08-29 | | Release date: | 2004-03-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the YDR533c S. cerevisiae protein, a class II member of the Hsp31 family
STRUCTURE, 12, 2004
|
|
3QYU
 
 | | Crystal structure of human cyclophilin D at 1.54 A resolution at room temperature | | Descriptor: | Peptidyl-prolyl cis-trans isomerase F | | Authors: | Colliandre, L, Gelin, M, Labesse, G, Guichou, J.-F. | | Deposit date: | 2011-03-04 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | In-plate protein crystallization, in situ ligand soaking and X-ray diffraction.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3RDC
 
 | | Human Cyclophilin D Complexed with an Inhibitor | | Descriptor: | Peptidyl-prolyl cis-trans isomerase F, mitochondrial, ethyl N-[(4-aminobenzyl)carbamoyl]glycinate | | Authors: | Colliandre, L, Ahmed-Belkacem, H, Bessin, Y, Pawlotsky, J.M, Guichou, J.F. | | Deposit date: | 2011-04-01 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Combining 'dry' co-crystallization and in situ diffraction to facilitate ligand screening by X-ray crystallography.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2RL3
 
 | | Crystal structure of the OXA-10 W154H mutant at pH 7 | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase PSE-2, GLYCEROL, ... | | Authors: | Vercheval, L, Kerff, F, Herman, R, Sauvage, E, Guiet, R, Charlier, P, Frere, J.-M, Galleni, M. | | Deposit date: | 2007-10-18 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Critical role of tryptophan 154 for the activity and stability of class D beta-lactamases.
Biochemistry, 48, 2009
|
|
2WGV
 
 | | Crystal structure of the OXA-10 V117T mutant at pH 6.5 inhibited by a chloride ion | | Descriptor: | BETA-LACTAMASE OXA-10, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Vercheval, L, Kerff, F, Bauvois, C, Sauvage, E, Guiet, R, Charlier, P, Galleni, M. | | Deposit date: | 2009-04-27 | | Release date: | 2010-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three Factors that Modulate the Activity of Class D Beta-Lactamases and Interfere with the Post- Translational Carboxylation of Lys70.
Biochem.J., 432, 2010
|
|
2WGW
 
 | | Crystal structure of the OXA-10 V117T mutant at pH 8.0 | | Descriptor: | BETA-LACTAMASE OXA-10, GLYCEROL, SULFATE ION | | Authors: | Vercheval, L, Kerff, F, Bauvois, C, Sauvage, E, Guiet, R, Charlier, P, Galleni, M. | | Deposit date: | 2009-04-27 | | Release date: | 2010-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three Factors that Modulate the Activity of Class D Beta-Lactamases and Interfere with the Post-Translational Carboxylation of Lys70.
Biochem.J., 432, 2010
|
|
2WGI
 
 | | Crystal structure of the acyl-enzyme OXA-10 W154A-benzylpenicillin at pH 6 | | Descriptor: | BETA-LACTAMASE OXA-10, GLYCEROL, OPEN FORM - PENICILLIN G | | Authors: | Vercheval, L, Falzone, C, Sauvage, E, Herman, R, Charlier, P, Galleni, M, Kerff, F. | | Deposit date: | 2009-04-20 | | Release date: | 2009-11-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Critical Role of Tryptophan 154 for the Activity and Stability of Class D Beta-Lactamases.
Biochemistry, 48, 2009
|
|
2WKI
 
 | | Crystal structure of the OXA-10 K70C mutant at pH 7.0 | | Descriptor: | 1,2-ETHANEDIOL, BETA-LACTAMASE OXA-10, GLYCEROL, ... | | Authors: | Vercheval, L, Bauvois, C, Kerff, F, Sauvage, E, Guiet, R, Charlier, P, Galleni, M. | | Deposit date: | 2009-06-11 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three Factors that Modulate the Activity of Class D Beta-Lactamases and Interfere with the Post-Translational Carboxylation of Lys70.
Biochem.J., 432, 2010
|
|
2X02
 
 | | CRYSTAL STRUCTURE OF THE CLASS D BETA-LACTAMASE OXA-10 AT 1.35 A RESOLUTION | | Descriptor: | BETA-LACTAMASE OXA-10, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Vercheval, L, Kerff, F, Sauvage, E, Herman, R, Galleni, M, Charlier, P. | | Deposit date: | 2009-12-04 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Impact of the Carboxylated Lysine on the Acylation and Deacylation Step in Class D Beta-Lactamase
To be Published
|
|
2X01
 
 | | CRYSTAL STRUCTURE OF THE OXA-10 S67A MUTANT AT PH 7 | | Descriptor: | BETA-LACTAMASE OXA-10, DIMETHYL SULFOXIDE, SULFATE ION, ... | | Authors: | Vercheval, L, Kerff, F, Bauvois, C, Sauvage, E, Guiet, R, Galleni, M, Charlier, P. | | Deposit date: | 2009-12-04 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evidence of Chloride Inhibition and Impact of the Hydrophobic Core on the Lysine Carboxylated in Class D Beta-Lactamase
To be Published
|
|
